 |
 |
 |
 |
 Multi-User Multi-Display Environment Multi-User Multi-Display Environment
We have adapted an open-source single-user, single-display molecular visualization application for use in a multi-display, multi-user
environment (figure left). Jmol, a popular, open-source Java applet for viewing PDB (protein databank) files, is modified in such a manner that allows synchronized coordinated views of the same molecule to be
displayed in a multi-display workspace. Each display in the workspace is driven by a separate PC, and coordinated views are achieved through the passing of RasMol script
commands over the network.  The environment includes a tabletop display capable of sensing touch-input, two large vertical displays, and a TabletPC. The presentation of large
molecules is adapted to best take advantage of the different qualities of each display, and a set of interaction techniques that allow groups working in this environment to better
collaborate are also presented. The environment includes a tabletop display capable of sensing touch-input, two large vertical displays, and a TabletPC. The presentation of large
molecules is adapted to best take advantage of the different qualities of each display, and a set of interaction techniques that allow groups working in this environment to better
collaborate are also presented.
Use of the Nintendo Wiimote
The Nintendo Wiimote is an inexpensive high degree-of-freedom wireless (bluetooth) input device. This suggests that the Wiimote might find use in a wide range of visualization applications including the manipulation of complex scientific data. We developed a series of inexpensive Wiimote-based interfaces for molecular visualization. These applications provide features previously only found in immersive environments to the low-cost desktop environment. We found that Wiimote-based molecular manipulation is particularly useful for presentations. When delivering a seminar, the speaker is able to interact directly with the projected image (rather than alternating between the presentation computer and the screen). In a classroom setting, an instructor can pass the Wiimote around the room evoking increased participation and exciting the students thought the "wow" factor.
Molecular Manipulation
 The Wiimote (figure right) provides a very natural interface for grabbing, rotating, and interacting with three-dimensional objects such as proteins. We extended the Jmol open-source Java applet for molecular visualization to incorporate Wiimote support. In our application, the Wiimote can function in several modes. Normal mouse mode allows the user to interact with Jmol in exactly the same
The Wiimote (figure right) provides a very natural interface for grabbing, rotating, and interacting with three-dimensional objects such as proteins. We extended the Jmol open-source Java applet for molecular visualization to incorporate Wiimote support. In our application, the Wiimote can function in several modes. Normal mouse mode allows the user to interact with Jmol in exactly the same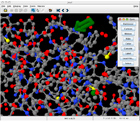 manner as with a standard two button mouse - this represents the 'easiest' mode for the user to learn. In the more advanced Cursor mode, the Wiimote controls a three-dimensional cursor (green arrow in figure left) in the molecular visualization window. The use of a three-dimensional pointer alleviates referencing ambiguities that can arise with a two-dimensional pointer. In our application, the user can 'stamp out' an arrow to mark a location. The stamped arrow is positioned in molecular coordinates and therefore rotates appropriately with the molecule. In both modes, the Wiimote can be used for controlling rotation, translation, and zoom. Navigation is complemented with a context sensitive option panel with large buttons (figure left). The large buttons make it easier to target visualization option widgets. manner as with a standard two button mouse - this represents the 'easiest' mode for the user to learn. In the more advanced Cursor mode, the Wiimote controls a three-dimensional cursor (green arrow in figure left) in the molecular visualization window. The use of a three-dimensional pointer alleviates referencing ambiguities that can arise with a two-dimensional pointer. In our application, the user can 'stamp out' an arrow to mark a location. The stamped arrow is positioned in molecular coordinates and therefore rotates appropriately with the molecule. In both modes, the Wiimote can be used for controlling rotation, translation, and zoom. Navigation is complemented with a context sensitive option panel with large buttons (figure left). The large buttons make it easier to target visualization option widgets.
Head-Tracking for Visualization
The notion of manipulating a three-dimensional image in response to the tracking of a user's head position has been an integral part of virtual-reality environments for over ten years. By rotating the visualization in a direction exactly opposite to the user's head motion a three-dimensional effect is achieved. In essence, the user's visual system is tricked into believing that he or she is looking around the object. Most recently, Johnny Lee, a graduate student at CMU, showed that this effect can be achieved utilizing Wiimote based tracking. In this system, the user is equipped with a pair of infrared emitting glasses; the user's head is then tracked using the Wiimote's infrared camera. This simple solution works surprisingly well. We adapted this technique for molecular visualization using the open-source application PyMol.
A short demonstration video is now available. We adapted an open source molecular visualization package (PyMol) to integrate two forms of head tracking (wiimote IR based and webcam based). In the demonstration video, we first illustrate active tracking using the wiimote and IR-emitting eyewear and then show our passive tracking using only Apple's built-in webcam. Click HERE.
Our software for Wiimote-based molecular visualization is available HERE (released as open source in April 2008). We are currently working with the developers of PyMol to make the software as easy to use as possible.

|
 |
 |
 |
 |
|
|
|