 |
 |
 |
 |

Despite the widespread use of macromolecular structures in unraveling the biochemical basis of life, state-of-the-art macromolecular structure determination via the two primary experimental methodologies, X-ray Crystallography and Nuclear Magnetic Resonance spectroscopy, remains a long, difficult, and expensive endeavour. Structure determination by X-ray Crystallography (XRC) involves the diffraction of high-energy X-rays through a pure protein crystal. The diffraction pattern observed (figure up-left) corresponds to a Fourier transform of the crystal's electron density and can be inverse transformed to recover the crystal’s molecular structure. Unfortunately, experimental diffraction data suffers from an information gap, the diffraction data contains only the magnitudes (or structure factors) and not the phases of each term of the Fourier transform. As a result, we do not have sufficient information to invert the function. The critical step of recovering diffraction phases (solving the phase problem) typically requires significant effort.
In the future, macromolecular structure determination will be an automated turn-key operation, and should be fast, inexpensive, and high-yield. My research focuses on developing computationally precise and efficient algorithms to expedite the process of protein structure determination. We focus on computational methods for addressing the phase problem of X-ray Crystallography.
For the Non-Biologist: The mantra that protein function is defined by protein structure drives the current interest in experimental protein structure determination. Knowledge of protein structure and function is helpful both in the analysis of natural wildtype proteins as well as mutant proteins implicated in disease. State-of-the-art methods for experimental protein structure determination are expensive ($120,000 per structure) and time consuming (~12 months per structure). Algorithms capable of efficiently processing experimental data and those involved in optimized experiment planning have and will continue to make significant impact.
|
 |
 |
 |
 |
 |
 |
 |
 |
 Traditional approaches to solving the phase problem fall into two classes, experimental methods (such as the use of heavy atoms (MIR) and x-ray phase-shifting (MAD)) and computational methods (such as homology modeling (Molecular Replacement - MR) and a brute force-type search (Direct Methods)). Both MIR and MAD require additional, potentially hazardous, wetlab experiments, adding additional cost and time to the structure determination effort. Molecular Replacement and Direct methods have the benefit of being purely computational. However, direct methods have limited application as they require high resolution data and small molecule size (up to 1000 atoms); they are therefore less useful for solving protein structures. Therefore, MR is typically the initial phasing method of choice.
Traditional approaches to solving the phase problem fall into two classes, experimental methods (such as the use of heavy atoms (MIR) and x-ray phase-shifting (MAD)) and computational methods (such as homology modeling (Molecular Replacement - MR) and a brute force-type search (Direct Methods)). Both MIR and MAD require additional, potentially hazardous, wetlab experiments, adding additional cost and time to the structure determination effort. Molecular Replacement and Direct methods have the benefit of being purely computational. However, direct methods have limited application as they require high resolution data and small molecule size (up to 1000 atoms); they are therefore less useful for solving protein structures. Therefore, MR is typically the initial phasing method of choice.
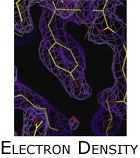
Methods, such as molecular replacement, that do not require additional wetlab experiments bridge the information gap to recover diffraction phases by making use of prior structural knowledge. In the case of molecular replacement, protein specific knowledge, in the form of an homologous model, bootstraps phase refinement. Because an homologous structure is used to bootstrap phase refinement, MR is generally not useful when determining the structures of proteins with novel folds. Unfortunately, it is exactly these novel structures that are of significant interest to structural biologists. We are currently extending molecular replacement  to function in the absence of a strong homologous model. This project presents as a combinatorial optimization problem in the presence of large and noisy datasets. We address these challenges by identifying mutually consistent data in the context of strong model priors. For example, our algorithm CRANS, employs algebraic subgroup identification to exploit non-crystallographic symmetry in molecular replacement. CRANS uses structural biochemical knowledge to compensate for low structural homology; and we used the algorithm to solve the structure of dihydrofolate reductase-thymidylate synthase from Cryptosporidium hominis (PDB: 1QZF) (figure left). to function in the absence of a strong homologous model. This project presents as a combinatorial optimization problem in the presence of large and noisy datasets. We address these challenges by identifying mutually consistent data in the context of strong model priors. For example, our algorithm CRANS, employs algebraic subgroup identification to exploit non-crystallographic symmetry in molecular replacement. CRANS uses structural biochemical knowledge to compensate for low structural homology; and we used the algorithm to solve the structure of dihydrofolate reductase-thymidylate synthase from Cryptosporidium hominis (PDB: 1QZF) (figure left).

|
 |
 |
 |
 |
|
|
|