 |
 |
 |
 |
Molecular procurement is a fundamental challenge for chemists working with novel molecules. One laborious technique for obtaining molecular samples is to extract them from a natural source (i.e., from a plant or animal source). Alternatively, a chemist can synthesize the desired molecule from commercially available starting reactants. To facilitate this process, a number of electronic chemical reaction databases are now available. These databases allow the user to query for single-step molecular transformations. A typical synthetic plan requires a sequence of approximately ten single step reactions to be chained together.
The CAOSP problem can be cast into the framework of a traditional AI planning problem, for example robot motion planning. These path planning algorithms utilize a search tree with internal nodes representing configurations or states and edges representing actions. A set of nodes are designated goal nodes; 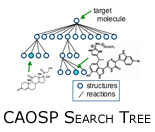 a path from the root node to a goal node represents a feasible path. In CAOSP, nodes represent molecules and edges represent chemical reactions capable of converting one molecule into another. a path from the root node to a goal node represents a feasible path. In CAOSP, nodes represent molecules and edges represent chemical reactions capable of converting one molecule into another.
Instead of performing a forward search, approaches to CAOSP typically gain computational leverage by employing a retrosynthetic search. A retrosynthetic plan starts with the target molecule and applies chemical reactions in reverse, generating a set of reactants from a product. Working backwards in this way, a search algorithm can stop when all required reactants are commercially available starting materials.
Typical chemical synthesis plans contain between 6 and 15 steps. Fortunately, this limits the depth of our search tree (i.e., we can limit the tree depth to ~15).  Unfortunately, our branching factor is enormous. For comparison, the game of chess is typically considered to have a branching factor of 35 - CAOSP has a branching factor between 500 and 10,000! On the bright side, CAOSP does not play against an adversary! Beyond the exponential nature of the search space, a second challenge relates to what we term reaction execution. A chemical reaction database is less a database of explicit chemical reactions and more a list of generalized reaction schemas. Reaction execution refers to predicting reaction products given a reaction schema and set of chemical reactants. The overabundance of special cases make reaction execution extremely difficult. Unfortunately, our branching factor is enormous. For comparison, the game of chess is typically considered to have a branching factor of 35 - CAOSP has a branching factor between 500 and 10,000! On the bright side, CAOSP does not play against an adversary! Beyond the exponential nature of the search space, a second challenge relates to what we term reaction execution. A chemical reaction database is less a database of explicit chemical reactions and more a list of generalized reaction schemas. Reaction execution refers to predicting reaction products given a reaction schema and set of chemical reactants. The overabundance of special cases make reaction execution extremely difficult.
We are currently collaborating with Simulated Biomolecular Systems author of ARChem and a leader in CAOSP. Check back to learn more.
 Modeling Stereochemistry: Many chemical reactions introduce or remove chirality; this must be properly modeled in a CAOSP algorithm. Modeling Stereochemistry: Many chemical reactions introduce or remove chirality; this must be properly modeled in a CAOSP algorithm. Robustness to Execution Ambiguity: Design of CAOSP algorithms that are robust to the uncertainty of reaction execution. Robustness to Execution Ambiguity: Design of CAOSP algorithms that are robust to the uncertainty of reaction execution. Domain Specific Search: Many modern advances in search have not found their way into CAOSP. We are interested in exploring how techniques such as A*, Tabu Search, and Gossip Protocols can be adapted to CAOSP using domain specific knowledge. Domain Specific Search: Many modern advances in search have not found their way into CAOSP. We are interested in exploring how techniques such as A*, Tabu Search, and Gossip Protocols can be adapted to CAOSP using domain specific knowledge. Molecular Synthetic Complexity: Efficient pruning of the search tree requires tight bounds on the synthetic complexity of molecular intermediates. Provable measures of synthetic complexity would allow more aggressive branch-and-bound search. Molecular Synthetic Complexity: Efficient pruning of the search tree requires tight bounds on the synthetic complexity of molecular intermediates. Provable measures of synthetic complexity would allow more aggressive branch-and-bound search. Structure-Based Drug Design: We are working to combine our research in CAOSP with our work in de novo Structure-Based Drug Design. Structure-Based Drug Design: We are working to combine our research in CAOSP with our work in de novo Structure-Based Drug Design.

|
 |
 |
 |
 |
|
|
|