SignalSpider
Probabilistic Modeling and Pattern Discovery on Multiple Normalized ChIP-Seq Signal Profiles
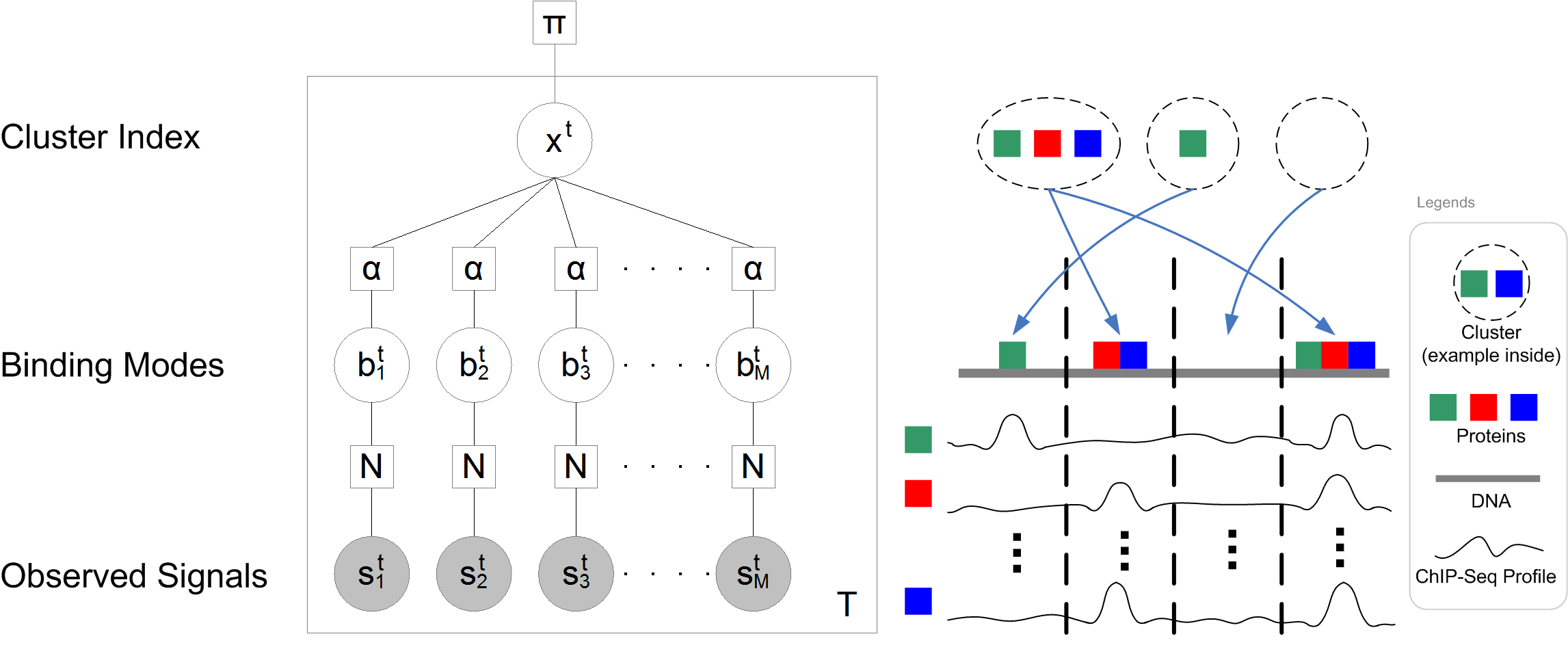
Overview
The technology of Chromatin immunoprecipitation (ChIP) followed by high-throughput sequencing (ChIP-Seq) measures the genome-wide occupancy of DNA-binding proteins in vivo. On the other hand, it is well-known that different combinations of DNA-binding protein occupancies may result in a gene being regulated in different tissues or at different developmental stages. To fully understand a gene's function, we propose a probabilistic model for deciphering the combinatorial binding of DNA-binding proteins. The model (SignalSpider) aims at modeling and extracting patterns from multiple ChIP-Seq profiles. We have built the SignalSpider models on the normalized ChIP-Seq profiles from Wiggler. With the support of Gene Ontology (GO) enrichment analysis, evolutionary conservation, chromatin interaction enrichment analysis, and wet-lab studies, we found that the discovered patterns show meaningful biological insight in a genome-wide scale.
Downloads
win64 |
linux64 |
|
| MCR (Matlab Compiler Runtime) | ||
| SignalSpider Executables and Demo Dataset (zipped) |
Command Usage
SignalSpider inFilePath [outModelPath] [numOfCombinations] [numOfBindingModes] [threshold] [maxIterations] [replicates]
| Input Arguments: | |
| inFilePath | The input profile signal file path (example: testData.csv) |
| outModelPath | The output SignalSpider model file path (default: SSmodel.mat) |
| numOfCombinations | Number of Clusters (default: 2) |
| numOfBindingModes | Number of Binding Modes (default: 2) |
| threshold | The tolerance used for testing convergence (default: 0.0001) |
| maxIterations | Maximal number of EM iterations (default: 100) |
| replicates | Number of replicates (default: 10) |
| Output Files: | |
| A SignalSpider model file as specified in the input argument "outModelPath" (default: SSmodel.mat) | |
| Modeling analysis files with the input argument "outModelPath" as the prefix of filenames. | |
| Microsoft Windows 64-bits examples: | |
| C:\> SignalSpider <argument_list> | |
| C:\> SignalSpider testData.csv | |
| C:\> SignalSpider testData.csv simpleModel.mat | |
| C:\> SignalSpider testData.csv fancyModel.mat 10 3 0.00001 1000 100 | |
| Linux 64-bits examples: | |
| >./run_SignalSpider.sh <mcr_directory> <argument_list> | |
| >./run_SignalSpider.sh /mathworks/home/application/v80 testData.csv | |
| >./run_SignalSpider.sh /mathworks/home/application/v80 testData.csv simpleModel.mat | |
| >./run_SignalSpider.sh /mathworks/home/application/v80 testData.csv fancyModel.mat 10 3 0.00001 1000 100 | |
FAQ
What is MCR ?
MCR is Matlab Compiler Runtime. If your machine does not have Matlab, you need to install MCR to execute SignalSpider. MCR can be downloaded from the internet easily. In particular, we advise you to download the same version indicated in the "Downloads" section.
Is there any demo ?
By default, a small testing dataset (testData.csv) is zipped with the SignalSpider executables in the "Downloads" section. Once downloaded, you can simply change your current directory to it and type "SignalSpider testData.csv" to run a SignalSpider demo on the testing dataset (which has 2 co-associate protein profiles). After the run, you will see the MATLAB SignalSpider model file (default: SSmodel.mat) and the image files as follow:
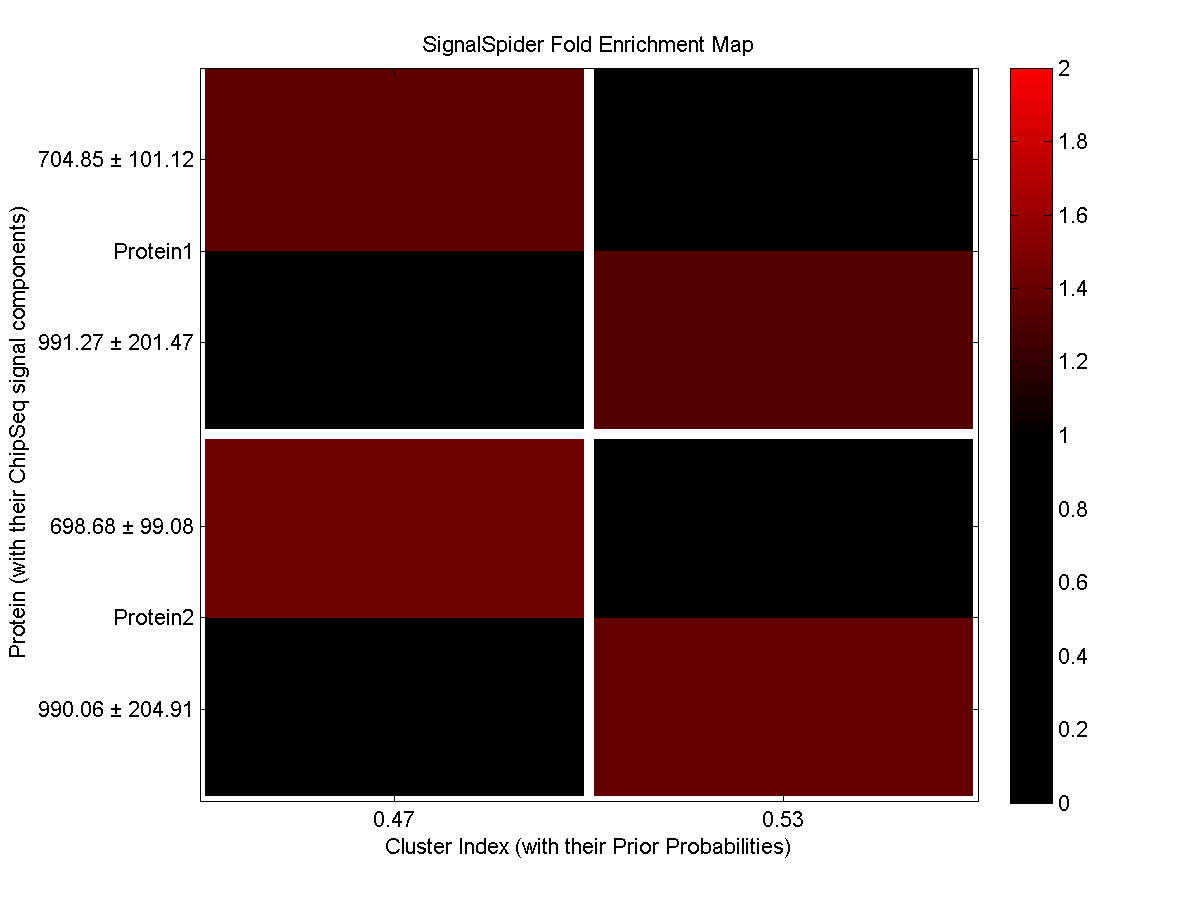 _
_
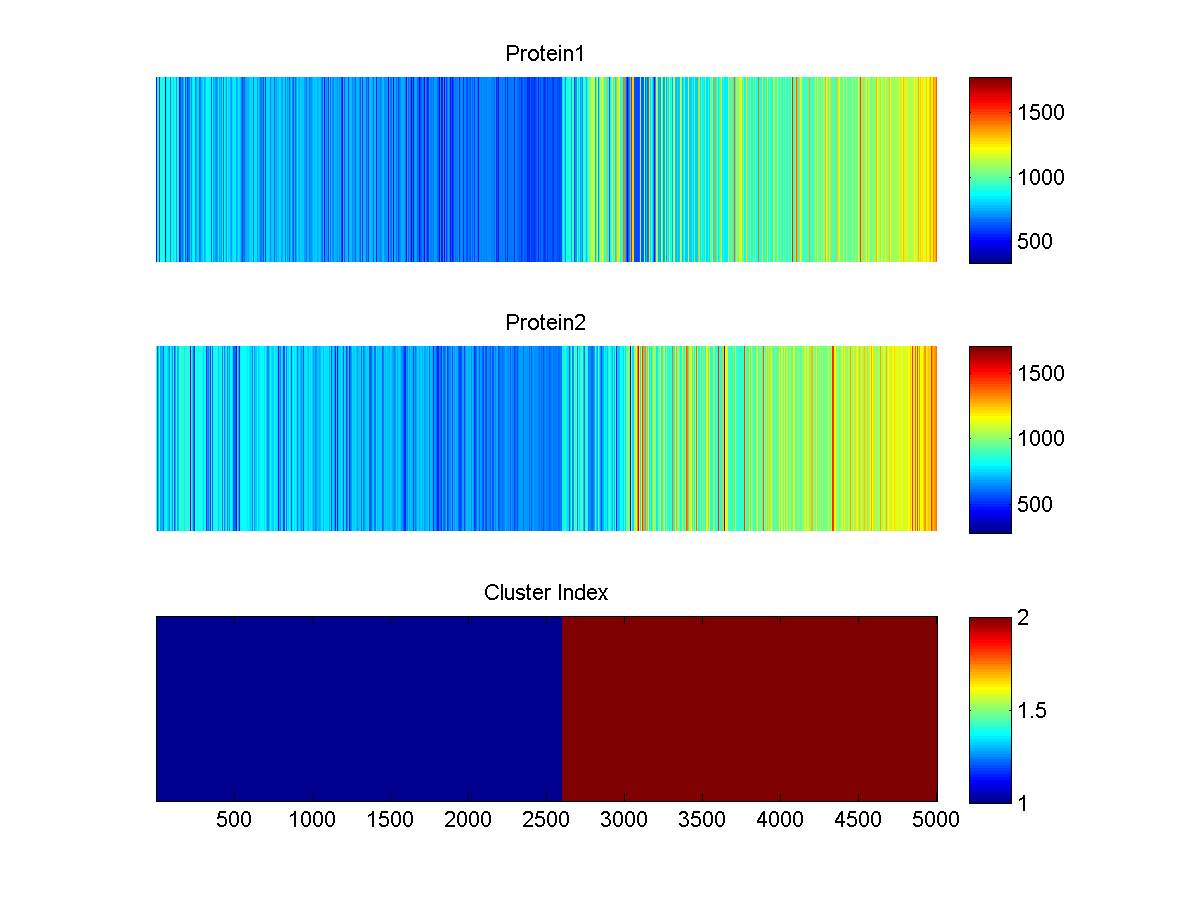 _
_
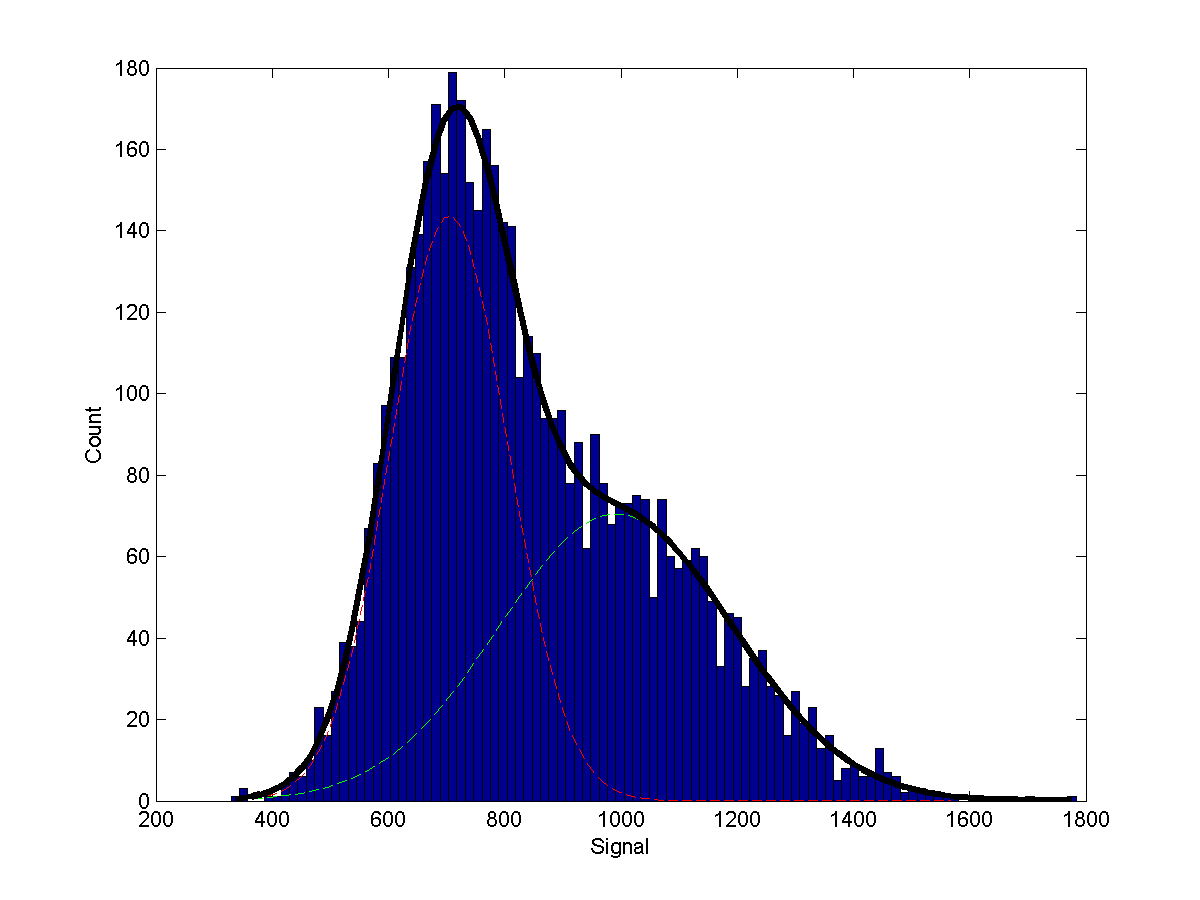 _
_
 _
_
More data ?
Public ChIP-Seq data can be accessed through ENCODE consortium and Gene Expression Omnibus (GEO). Details of Wiggler can be found here.
More questions ?
Please contact Ka-Chun Wong
Citation
© 2014 Ka-Chun Wong
Template design by Andreas Viklund