

- Our Vision: A healthier world.
- Our Mission: Accelerating science by creating a supercomputer empowered by a global community of volunteers.
- “WCG continues to support open-source and opendata research and helps reduce computational time to empower scientists to address the world’s most pressing questions at no cost to the researchers”. Dr. Igor Jurisica
- “By its very nature, World Community Grid is a collaborative effort involving individuals and institutions from around the world working towards innovative advancements in science that benefit humanity. Distributive is proud to be a part of this mission.” Dr. Dan Desjardins, co-founder & CEO, Distributive Corp., Kingston, Ontario, Canada
- “This work has demonstrated the importance of the integration of computational and experimental approaches, as well as the potential of large-scale collaborative networks to advance drug discovery projects for neglected diseases and emerging viruses, despite the lack of available direct antiviral activity and cytoprotective effect data, that reflects on the assertiveness of the computational predictions.” Dr. Carolina Horta Andrade; Open Zika project
- “With the help of volunteers, partners, and institutions, the WCG will continue to grow as the world's largest volunteer-driven supercomputer, enabling seemingly impossible scientific research to come to life”.

World Community Grid (WCG) is a public, high performance computing platform available for open science/open data research that benefits humanity. It is a global, heterogeneous, distributed cloud infrastructure that runs on secure BOINC platform developed at Berkeley. Over the 19+ years of its operation, 814,927 volunteers with more than 7.69 million devices contributed over 2.619 million CPU years towards research on cancer, tuberculosis, COVID-19, HIV/AIDS, microbiome immunity, understanding genomes and protein structure, developing new materials, clean energy, ensuring sustainable water, understanding weather patterns in Africa, and other globally important projects.
WCG diminished computational bottlenecks for open science and increased research breakthroughs by using scalable yet sustainable computing. It brings together volunteers and researchers at the intersection of computational biology, open science, open data, and citizen science - four trends that are transforming the way research is conducted. WCG has supported 31 research projects to date, and enabled seemingly impossible scientific research to come to life. WCG condenses computational time and enables scientists to answer the world’s most pressing questions. Our research partners have published over 186 peer-reviewed papers in scientific journals. WCG is the biggest volunteer computing initiative devoted to science benefiting humanity, and is as powerful as some of the world's fastest supercomputers.
WCG was one of the first to use GPUs to accelerate scientific computation on the Help Conquer Cancer project, and in 2013, it also became one of the first major volunteer computing initiatives to enable mobile computing on Android smartphones and tablets.
History
WCG began in 2004 as a philanthropic initiative of IBM Corporate Social Responsibility, the corporate social responsibility and philanthropy division of IBM. Through Corporate Social Responsibility, IBM donated its technology and talent to address some of the world's most pressing social and environmental issues. WCG has been pioneering and an award-winning program, which has been recognized internationally with awards, including the Computerworld Data+ Editors’ Choice Award, Business in the Community Coffey International Award, and the Asian Forum on Corporate Social Responsibility's Asian CSR Award.
In 2021, IBM transferred the World Community Grid assets to Jurisica Lab at Krembil Research Institute, University Health Network (UHN) in Toronto, Canada. While sharing the goals and principles of WCG, we aim to expand the mission of citizen science, youth outreach and integrative computational biology. UHN has Canada’s largest hospital-based research program, comprising three major teaching hospitals (Toronto Western Hospital, Toronto General Hospital, Princess Margaret Cancer Centre) and their research insititutes, Toronto Rehabilitation Institute, and The Michener Institute of Education.
Dr. Igor Jurisica’s research is connected to the Toronto Western Hospital and the hospital’s research arm, the Krembil Research Institute; a non-profit academic biomedical research institute. Research within Krembil is focused on the development of diagnostics, treatments and management strategies in the following three programmatic areas:
- 1) arthritis and musculoskeletal system disorders, e.g., osteoarthritis, rheumatoid arthritis, systemic lupus erythematosus, and ankylosing spondylitis;
- 2) brain and chronic neurological/neurosurgical disorders, e.g., concussion and traumatic brain injury, Alzheimer's and Parkinson's diseases, stroke, epilepsy, spinal cord injuries, dementia, pain and depression;
- 3) vision and ophthalmologic disorders, e.g., glaucoma, macular degeneration, and retinopathy.



Mapping Cancer Markers aims to identify markers associated with various types of cancer. This will help detect cancer earlier and design more personalized cancer care.

Scientists at Scripps Research are using World Community Grid to help search for potential treatments for COVID-19, and to build open-source tools to help address future pandemics quickly and early.

95% of agriculture in Africa depends on rainfall. This project uses massive computing power, data from The Weather Company, and other data to provide more accurate rainfall forecasts

Since 2009, World Community Grid has been part of the fight against childhood cancer. This project continues that work by expanding the search for better treatments to more types of childhood cancer.
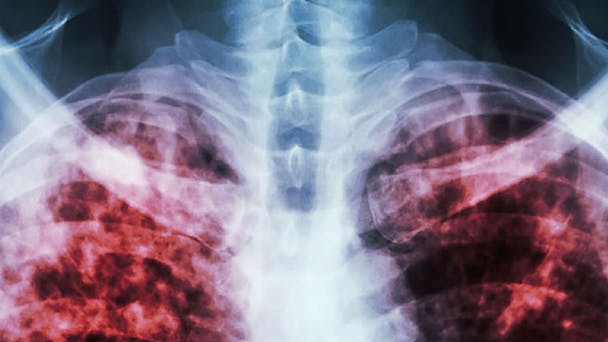
Tuberculosis killed 1.5 million people in 2014, making it one of the world's deadliest diseases. You can help researchers learn more about this disease and how to overcome it.

World Community Grid volunteers donated nearly 146,000 CPU years to the MIP. This effort enabled the team to predict the structure of almost 200,000 proteins, discover over 150 new protein shapes (folds), describe several previously unknown functions of protein structures, and nearly double the number of annotated proteins in the human gut microbiome.

From almost 404 million results generated by WCG, amounting to almost 93 thousand years of computation, 61 hits were prioritised for testing. Further tests identified 8 compounds as being able to protect the cells from death caused by the virus, while showing low cellular toxicity in liver and kidney derived cells.

The first phase of FightAIDS@Home made significant advances in HIV research. As the virus evolves, the research team used new analysis techniques to better identify promising anti-HIV drugs.

Ebola is a deadly virus that kills up to 90% of infected victims. WCG and the volunteers helped to find the most promising drug leads to fight the Ebola virus.

Uncovering Genome Mysteries expects to examine close to 200 million genes from a wide variety of life forms, such as seaweeds from Australian coastlines and microbes found in Amazon river samples.

The project studied the effects of human activity on a large watershed and gain deeper insights into what actions can lead to restoration, health and sustainability of this important water resource. The extensive computing power of WCG was used to perform millions of computer simulations to better understand the effects that result from a variety of human activity patterns in the Chesapeake Bay area.

The extensive computing power of WCG was used to perform computer simulations of the interactions between millions of chemical compounds and certain target proteins. This has helped find the most promising compounds that may lead to effective treatments for Schistosoma.

The aim of the project was to discover promising new drug candidates that could be developed into new drugs that cure drug resistant forms of malaria, by performing computer simulations of the interactions between millions of chemical compounds and certain target proteins, to predict their ability to eliminate malaria.

The mission of Drug Search for Leishmaniasis was to identify potential drug candidates that could possibly be developed into treatments for Leishmaniasis by running computer simulations of the interactions between millions of chemical compounds and certain target proteins.

The mission of Computing for Clean Water was to provide deeper insight on the molecular scale into the origins of the efficient flow of water through a novel class of filter materials. This insight will in turn guide future development of low-cost and more efficient water filters.

The mission of The Clean Energy Project was to find new materials for the next generation of solar cells and later, energy storage devices. By harnessing the immense power of WCG, researchers have calculated the electronic properties of hundreds of thousands of organic materials - thousands of times more than could ever be tested in a lab - and identified the most promising candidates for developing affordable solar energy technology.

The project's goals were to uncover novel drugs to cure dengue hemorrhagic fever, hepatitis C, West Nile encephalitis, and Yellow fever; diseases caused by the infection with viruses from the family Flaviviridae.

The project investigated protein-protein interactions for more than 2,200 proteins whose structures are known, with particular focus on those proteins that play a role in neuromuscular diseases. The database of information produced is helping researchers design molecules to inhibit or enhance binding of particular macromolecules, hopefully leading to better treatments for muscular dystrophy and other neuromuscular diseases.

The aimed to find new drugs that can stop the spread of an influenza infection in the body. The research specifically focused on the influenza strains that have become drug resistant as well as new strains that are appearing, accelerating the efforts to develop treatments that would be useful in managing seasonal influenza outbreaks, and future influenza epidemics and even pandemics.

The mission of the Help Fight Childhood Cancer project has been to find drugs that can disable three particular proteins associated with neuroblastoma, one of the most frequently occurring solid tumors in children. Identifying these drugs could potentially make the disease much more curable when combined with chemotherapy treatment.

The mission of the Clean Energy Project was to find new materials for the next generation of solar cells and later, energy storage devices.

The objective of this project was to predict the structure of proteins of major strains of rice, which in turn will help farmers breed better rice strains with higher crop yields, promote greater disease and pest resistance, and utilize a full range of bioavailable nutrients that can benefit people around the world, especially in regions where hunger is a critical concern.

The main goal of the project was to improve the results of protein X-ray crystallography in order to increase understanding of cancer and its treatment.

The mission of AfricanClimate@Home was to develop more accurate climate models of specific regions in Africa. This will serve as a basis for understanding how the climate will change in the future so that measures designed to alleviate the adverse effects of climate change can be implemented.

The mission of the project was to identify promising drug leads to combat the Dengue, Hepatitis C, West Nile, Yellow Fever, and other related viruses. The extensive computing power of World Community Grid was used to complete the structure-based drug discovery calculations required to identify these leads.

This project focused on investigating protein-protein, protein-DNA and protein-ligand interactions for the 10,000 proteins whose structures are known, with particular focus on those proteins that play a role in neuromuscular diseases. The database of information produced will help researchers design molecules to inhibit or enhance binding of particular macromolecules, hopefully leading to better treatments for muscular dystrophy and other neuromuscular diseases.

WThe project focused on comparing genomic information to improve the quality and interpretation of biological data and our understanding of biological systems, host-pathogen and environmental interactions. This information can play a critical role in the development of better drugs and vaccines, and improved diagnostic procedures.

The project was examining tissue microarrays to determine how to improve the treatment of cancer with earlier and more targeted diagnostic tools.

Human Proteome Folding Phase 2 (HPF2) continued where the first phase left off. The two main objectives of the project were to: 1) obtain higher resolution structures for specific human proteins and pathogen proteins and 2) further explore the limits of protein structure prediction by further developing Rosetta software structure prediction.

FightAIDS@Home focused on using computation methods to identify candidate drugs that have the right shape and chemical characteristics to block HIV protease.

The Human Proteome Folding project has generate data for predicting the shape of a very large number of human proteins. These predictions give scientists the clues they need to identify the biological functions of individual proteins within the human body. With an understanding of how each protein affects human health, scientists can develop new cures for human diseases such as cancer, HIV/AIDS, SARS, malaria, arthritis and many others .
- Cao, Wei, Jin Wang and Ming Ma. Carbon nanostructure based mechano-nanofluidics. Journal of Micromechanics and Microengineering (2018). DOI: 10.1088/1361-6439/aaa782.
- Ma, Ming, François Grey, Luming Shen, Michael Urbakh, Shuai Wu, Jefferson Zhe Liu, Yilun Liu and Quanshui Zheng. Water transport inside carbon nanotubes mediated by phonon-induced oscillating friction. Nature
- Nanotechnology (2015). DOI: 10.1038/nnano.2015.134.
- Ma, Ming D., Luming Shen, John Sheridan, Jefferson Zhe Liu, Chao Chen and Quanshui Zheng. Friction of water slipping in carbon nanotubes. Physical Review E (2011). DOI: 10.1103/PhysRevE.83.036316.
- Viswanathan, Usha, Suzanne M. Tomlinson, John M. Fonner, Stephen A. Mock and Stanley J. Watowich. Identification of a novel inhibitor of dengue virus protease through use of a virtual screening drug discovery Web portal. Journal of Chemical Information and Modeling (2014). DOI: 10.1021/ci500531r.
- Tomlinson, S. M., R. D. Malmstrom and S. J. Watowich. New Approaches to Structure-Based Discovery of Dengue Protease Inhibitors. Infectious Disorders - Drug Targets (2009). DOI: 10.2174/1871526510909030327.
- Ochoa, Rodrigo, Stanley J. Watowich, Andrés Flórez, Carol V. Mesa, Sara M. Robledo and Carlos Muskus. Drug search for leishmaniasis: a virtual screening approach by grid computing. Journal of Computer-Aided Molecular Design (2016). DOI: 10.1007/s10822-016-9921-4.
- Flórez, Andrés F., Stanley Watowich, Carlos Muskus, Andrés F. Flórez, Stanley Watowich and Carlos Muskus. Current Advances in Computational Strategies for Drug Discovery in Leishmaniasis. (2012).
- Sun, Q., Biswas, A., Vijayan, R.S.K. et al. Structure-based virtual screening workflow to identify antivirals targeting HIV-1 capsid. J Comput Aided Mol Des 36, 193–203 (2022). https://doi.org/10.1007/s10822-022-00446-5
- Goodsell, David S., Michel F. Sanner, Arthur J. Olson and Stefano Forli. The AutoDock suite at 30. Protein Science (2021). DOI: 10.1002/pro.3934.
- Craveur, Pierrick, Anna T. Gres, Karen A. Kirby et al. Novel Intersubunit Interaction Critical for HIV-1 Core Assembly Defines a Potentially Targetable Inhibitor Binding Pocket. mBio (2019). DOI: 10.1128/mBio.02858-18.
- Xia, Junchao, William Flynn, Emilio Gallicchio, Keith Uplinger, Jonathan D. Armstrong, Stefano Forli, Arthur J. Olson and Ronald M. Levy. Massive-Scale Binding Free Energy Simulations of HIV Integrase Complexes Using Asynchronous Replica Exchange Framework Implemented on the IBM WCG Distributed Network. Journal of Chemical Information and Modeling (2019). DOI: 10.1021/acs.jcim.8b00817.
- Forli, Stefano and Arthur J. Olson. Computational Challenges of Structure-Based Approaches Applied to HIV. The Future of HIV-1 Therapeutics (2015).
- Xia, Junchao, William F. Flynn, Emilio Gallicchio, Bin W. Zhang, Peng He, Zhiqiang Tan and Ronald M. Levy. Large-scale asynchronous and distributed multidimensional replica exchange molecular simulations and efficiency analysis. Journal of Computational Chemistry (2015). DOI: 10.1002/jcc.23996.
- Gallicchio, Emilio, Junchao Xia, William F. Flynn, Baofeng Zhang, Sade Samlalsingh, Ahmet Mentes and Ronald M. Levy. Asynchronous replica exchange software for grid and heterogeneous computing. Computer Physics Communications (2015). DOI: 10.1016/j.cpc.2015.06.010.
- Perryman, Alexander L., Daniel N. Santiago, Stefano Forli, Diogo Santos-Martins and Arthur J. Olson. Virtual screening with AutoDock Vina and the common pharmacophore engine of a low diversity library of fragments and hits against the three allosteric sites of HIV integrase: participation in the SAMPL4 protein–ligand binding challenge. Journal of Computer-Aided Molecular Design (2014). DOI: 10.1007/s10822-014-9709-3.
- Perryman, Alexander L., Qing Zhang, Holly H. Soutter, Robin Rosenfeld, Duncan E. McRee, Arthur J. Olson, John E. Elder and C. David Stout. Fragment-Based Screen against HIV Protease. Chemical Biology & Drug Design (2010). DOI: 10.1111/j.1747-0285.2009.00943.x.
- Perryman, Alex L., Stefano Forli, Garrett M. Morris et al. A Dynamic Model of HIV Integrase Inhibition and Drug Resistance. Journal of Molecular Biology (2010). DOI: 10.1016/j.jmb.2010.01.033.
- Cosconati, Sandro, Stefano Forli, Alex L Perryman, Rodney Harris, David S Goodsell and Arthur J Olson. Virtual Screening with AutoDock: Theory and Practice. Expert Opinion on Drug Discovery (2010). DOI: 10.1517/17460441.2010.484460.
- Morris, Garrett M., Ruth Huey, William Lindstrom, Michel F. Sanner, Richard K. Belew, David S. Goodsell and Arthur J. Olson. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry (2009). DOI: 10.1002/jcc.21256.
- Chang, Max W., William Lindstrom, Arthur J. Olson and Richard K. Belew. Analysis of HIV Wild-Type and Mutant Structures via in Silico Docking against Diverse Ligand Libraries. Journal of Chemical Information and Modeling (2007). DOI: 10.1021/ci700044s.
- Perryman, Alexander L., Weixuan Yu, Xin Wang et al. A Virtual Screen Discovers Novel, Fragment-Sized Inhibitors of Mycobacterium tuberculosis InhA. Journal of Chemical Information and Modeling (2015). DOI: 10.1021/ci500672v.
- Lifschitz, Sérgio, Carlos Juliano M. Viana, Cristian Tristão, Marcos Catanho, Wim M. Degrave, Antonio Basílio de Miranda, Márcia Bezerra and Thomas D. Otto. Design and Implementation of ProteinWorldDB. Advances in Bioinformatics and Computational Biology (2012).
- Otto, Thomas Dan, Marcos Catanho, Cristian Tristão et al. ProteinWorldDB: querying radical pairwise alignments among protein sets from complete genomes. Bioinformatics (2010). DOI: 10.1093/bioinformatics/btq011.
- Kotseruba, Yulia, Christian A. Cumbaa and Igor Jurisica. High-throughput protein crystallization on the World Community Grid and the GPU. Journal of Physics: Conference Series (2012). DOI: 10.1088/1742-6596/341/1/012027.
- Cumbaa, Christian A. and Igor Jurisica. Protein crystallization analysis on the World Community Grid. Journal of Structural and Functional Genomics (2010). DOI: 10.1007/s10969-009-9076-9.
- Snell, Edward H., Angela M. Lauricella, Stephen A. Potter et al. Establishing a training set through the visual analysis of crystallization trials. Part II: crystal examples. Acta Crystallographica Section D: Biological Crystallography (2008). DOI: 10.1107/S0907444908028059.
- Snell, Edward H., Joseph R. Luft, Stephen A. Potter et al. Establishing a training set through the visual analysis of crystallization trials. Part I: 150000 images. Acta Crystallographica Section D: Biological Crystallography (2008). DOI: 10.1107/S0907444908028047.
- Dequeker, Chloé, Elodie Laine and Alessandra Carbone. Decrypting protein surfaces by combining evolution, geometry, and molecular docking. Proteins: Structure, Function, and Bioinformatics (2019). DOI: 10.1002/prot.25757.
- Lagarde, Nathalie, Alessandra Carbone and Sophie Sacquin-Mora. Hidden partners: Using cross-docking calculations to predict binding sites for proteins with multiple interactions. Proteins: Structure, Function, and Bioinformatics (2018). DOI: 10.1002/prot.25506.
- Laine, Elodie and Alessandra Carbone. Protein social behavior makes a stronger signal for partner identification than surface geometry. Proteins: Structure, Function, and Bioinformatics (2017). DOI: 10.1002/prot.25206.
- Vamparys, Lydie, Benoist Laurent, Alessandra Carbone and Sophie Sacquin-Mora. Great interactions: How binding incorrect partners can teach us about protein recognition and function. Proteins: Structure, Function, and Bioinformatics (2016). DOI: 10.1002/prot.25086.
- Lopes, Anne, Sophie Sacquin-Mora, Viktoriya Dimitrova, Elodie Laine, Yann Ponty and Alessandra Carbone. Protein-Protein Interactions in a Crowded Environment: An Analysis via Cross-Docking Simulations and Evolutionary Information. PLOS Computational Biology (2013). DOI: 10.1371/journal.pcbi.1003369.
- Bertis, Viktors, Raphaël Bolze, Frédéric Desprez and Kevin Reed. From Dedicated Grid to Volunteer Grid: Large Scale Execution of a Bioinformatics Application. Journal of Grid Computing (2009). DOI: 10.1007/s10723-009-9130-7.
- Engelen, Stefan, Ladislas A. Trojan, Sophie Sacquin-Mora, Richard Lavery and Alessandra Carbone. Joint Evolutionary Trees: A Large-Scale Method To Predict Protein Interfaces Based on Sequence Sampling. PLOS Computational Biology (2009). DOI: 10.1371/journal.pcbi.1000267.
- Sacquin-Mora, Sophie, Alessandra Carbone and Richard Lavery. Identification of Protein Interaction Partners and Protein–Protein Interaction Sites. Journal of Molecular Biology (2008). DOI: 10.1016/j.jmb.2008.08.002.
- Foran, David J, Lin Yang, Wenjin Chen et al. ImageMiner: a software system for comparative analysis of tissue microarrays using content-based image retrieval, high-performance computing, and grid technology. Journal of the American Medical Informatics Association (2011). DOI: 10.1136/amiajnl-2011-000170.
- Wang, Fusheng. Grid-Enabled, High-performance Microscopy Image Analysis. (2010).
- Lin Yang, Wenjin Chen, P. Meer, G. Salaru, L.A. Goodell, V. Berstis and D.J. Foran. Virtual Microscopy and Grid-Enabled Decision Support for Large-Scale Analysis of Imaged Pathology Specimens. IEEE Transactions on Information Technology in Biomedicine (2009). DOI: 10.1109/TITB.2009.2020159.
- Lin Yang, O. Tuzel, Wenjin Chen, P. Meer, G. Salaru, L.A. Goodell and D.J. Foran. PathMiner: A Web-Based Tool for Computer-Assisted Diagnostics in Pathology. IEEE Transactions on Information Technology in Biomedicine (2009). DOI: 10.1109/TITB.2008.2008801.
- DiPaola, Robert S., Dmitri Dvorzhinski, Anu Thalasila et al. Therapeutic starvation and autophagy in prostate cancer: A new paradigm for targeting metabolism in cancer therapy. The Prostate (2008). DOI: 10.1002/pros.20837.
- Fukuda, Mayu, Atsushi Takatori, Yohko Nakamura, Akiko Suganami, Tyuji Hoshino, Yutaka Tamura and Akira Nakagawara. Effects of novel small compounds targeting TrkB on neuronal cell survival and depression-like behavior. Neurochemistry International (2016). DOI: 10.1016/j.neuint.2016.04.017.
- Nakamura, Yohko, Akiko Suganami, Mayu Fukuda et al. Identification of novel candidate compounds targeting TrkB to induce apoptosis in neuroblastoma. Cancer Medicine (2014). DOI: 10.1002/cam4.175.
- Groenewald, Wilma, Ricardo A. Parra-Cruz, Christof M. Jäger and Anna K. Croft. Revealing solvent-dependent folding behavior of mycolic acids from Mycobacterium tuberculosis by advanced simulation analysis. Journal of Molecular Modeling (2019). DOI: 10.1007/s00894-019-3943-5.
- Baltz, Alexander G., Mathias Munschauer, Björn Schwanhäusser et al. The mRNA-Bound Proteome and Its Global Occupancy Profile on Protein-Coding Transcripts. Molecular Cell (2012). DOI: 10.1016/j.molcel.2012.05.021.
- Pentony, M. M., P. Winters, D. Penfold-Brown, K. Drew, A. Narechania, R. DeSalle, R. Bonneau and M. D. Purugganan. The Plant Proteome Folding Project: Structure and Positive Selection in Plant Protein Families. Genome Biology and Evolution (2012). DOI: 10.1093/gbe/evs015.
- Drew, Kevin, Patrick Winters, Glenn L. Butterfoss et al. The Proteome Folding Project: Proteome-scale prediction of structure and function. Genome Research (2011). DOI: 10.1101/gr.121475.111.
- Boxem, Mike, Zoltan Maliga, Niels Klitgord et al. A Protein Domain-Based Interactome Network for C. elegans Early Embryogenesis. Cell (2008). DOI: 10.1016/j.cell.2008.07.009.
- Bonneau, Richard, Marc T. Facciotti, David J. Reiss et al. A Predictive Model for Transcriptional Control of Physiology in a Free Living Cell. Cell (2007). DOI: 10.1016/j.cell.2007.10.053.
- Malmström, Lars, Michael Riffle, Charlie E. M. Strauss, Dylan Chivian, Trisha N. Davis, Richard Bonneau and David Baker. Superfamily assignments for the yeast proteome through integration of structure prediction with the gene ontology. PLoS biology (2007). DOI: 10.1371/journal.pbio.0050076.
- Andersen-Nissen, Erica, Kelly D. Smith, Richard Bonneau, Roland K. Strong and Alan Aderem. A conserved surface on Toll-like receptor 5 recognizes bacterial flagellin. Journal of Experimental Medicine (2007). DOI: 10.1084/jem.20061400.
- Avila-Campillo, Iliana, Kevin Drew, John Lin, David J. Reiss and Richard Bonneau. BioNetBuilder: automatic integration of biological networks. Bioinformatics (2007). DOI: 10.1093/bioinformatics/btl604.
- vMalmström, Lars. Genome-wide structural and functional protein characterization by ab initio protein structure prediction. Report / Department of Electrical Measurements. Lund Institute of Technology (2005).
- Samarasinghe KW, Kotlyar M, Vallet SD, Hayes C, Naba A, Jurisica I, Lisacek F, Ricard-Blum S. MatrixDB 2024: an increased coverage of extracellular matrix interactions, a new Network Explorer and a new web interface. Nucleic Acids Res. 53(D1):D1677-D1682, 2025.
- Sigalotti L, Frezza AM, Sbaraglia M, Del Savio E, Baldazzi D, Valenti B, Bellan E, De Benedictis I, Doni M, Gambarotti M, Vincenzi B, Brunello A, Baldi GG, Palmerini E, Pasquali S, Ciuffetti ME, Varano V, Cappello F, Appolloni V, Pastrello C, Jurisica I, Gronchi A, Stacchiotti S, Casali PG, Dei Tos AP, Maestro R. Proximal and classic epithelioid sarcomas are distinct molecular entities defined by MYC/GATA3 and SOX17/endothelial markers, respectively. Mod Pathol, 38(1):100647, 2024.
- Maier A, Hartung M, Abovsky M, Adamowicz K, Bader GD, Baier S, Blumenthal DB, Chen J, Elkjaer ML, Garcia-Hernandez C, Helmy M, Hoffmann M, Jurisica I, Kotlyar M, Lazareva O, Levi H, List M, Lobentanzer S, Loscalzo J, Malod-Dognin N, Manz Q, Matschinske J, Mee M, Oubounyt M, Pastrello C, Pico AR, Pillich RT, Poschenrieder JM, Pratt D, Pržulj N, Sadegh S, Saez-Rodriguez J, Sarkar S, Shaked G, Shamir R, Trummer N, Turhan U, Wang RS, Zolotareva O, Baumbach J. Drugst.One - a plug-and-play solution for online systems medicine and network-based drug repurposing. Nucleic Acids Res, 52(W1):W481-W488, 2024.
- Pastrello C, Kotlyar M, Abovsky M, Lu R, Jurisica I. PathDIP 5: improving coverage and making enrichment analysis more biologically meaningful. Nucleic Acids Res, 52(D1):D663-D671, 2024.
- Wong SWH, Pastrello C, Kotlyar M, Faloutsos C, Jurisica I. USNAP: fast unique dense region detection and its application to lung cancer. Bioinformatics. 39(8):btad477, 2023.
- van Gogh M, Glaus Garzon JF, Sahin D, Knopfova L, Benes P, Boyman O, Jurisica I, Borsig L. Tumor cell-intrinsic c-Myb upregulation stimulates antitumor immunity in a murine colorectal cancer model, Cancer Immunol Res, CIR-22-0912, 2023. doi: 10.1158/2326-6066.CIR-22-0912.
- D'Angelo E, Pastrello C, Biccari A, Marangio A, Sensi F, Crotti S, Fassan M, Jurisica I, Pucciarelli S, Agostini M. An integrated multiomics analysis of rectal cancer patients identified POU2F3 as a putative druggable target and entinostat as a cytotoxic enhancer of 5-fluorouracil, Int J Cancer, 153(2):437-449, 2023.
- Houschild AC, Pastrello C, Ekaputeri GKA, Bethune-Waddell D, Abovsky M, Ahmed Z, Kotlyar M Lu R, Jurisica I. MirDIP 5.2: tissue context annotation and novel microRNA curation, Nuncl Acids Res, 51(D1):D217-D225, 2023.
- Pathmanathan S, Yao Z, Coelho P, Valla R, Drecun L, Benz C, Snider J, Saraon P, Grozavu I, Kotlyar M, Jurisica I, Park M, Stagljar I. B cell linker protein (BLNK) is a regulator of Met receptor signaling and trafficking in non-small cell lung cancer, iScience, 25(11):105419, 2022.
- Agapito G, Niu Y, Pastrello C, Jurisica I. Pathway integration and annotation: building a puzzle with non-matching pieces and no picture to follow, Briefings in Bioinformatics, 23(5):bbac368, 2022.
- Pastrello C, Abovsky M, Lu R, Ahmed Z, Kotlyar M, Veillette C, Jurisica I, Osteoarthritis Data Integration Portal (OsteoDIP): A web-based gene and non-coding RNA expression database, Osteoarthritis and Cartilage Open, 4(1): 100237, 2022.
- Tokar T, Pastrello C, Abovsky M, Rahmati S, Jurisica I. miRAnno-network-based functional microRNA annotation, Bioinformatics, 38(2): 592-593, 2022.
- Kotlyar, M., Pastrello, C., Ahmed, Z., Chee, J., Varyova, Z., Jurisica, I., IID 2021: Towards context-specific protein interaction analyses by increased coverage, enhanced annotation and enrichment analysis. Nucl Acids Res, 50(D1):D640-D647, 2022. doi: 10.1093/nar/gkab1034.
- Bhat M, Pasini E, Pastrello C, Angeli M, Baciu C, Abovsky M, Coffee A, Adeyi O, Kotlyar M, Jurisica I, Estrogen Receptor 1 Inhibition of Wnt/Beta-catenin Signaling Contributes to Sex Differences in Hepatocarcinogenesis, Frontiers in Oncology, section Molecular and Cellular Oncology, 11:777834. doi: 10.3389/fonc.2021.777834. eCollection 2021.
- Cohn DE, Barros-Filho MC, Minatel BC, Pewarchuk ME, Marshall EA, Vucic EA, Sage AP, Telkar N, Stewart GL, Jurisica I, Reis PP, Robinson WP, Lam WL. Share, Reactivation of Multiple Fetal miRNAs in Lung Adenocarcinoma, Cancers, 13(11):2686, 2021.
- Bhat, M. Elisa Pasini, Chiara Pastrello, Sarah Rahmati, Marc Angeli, Max Kotlyar, Anand Ghanekar and Igor Jurisica, Integrative Analysis of Layers of Data in Hepatocellular Carcinoma Reveals Pathway Dependencies, World J Hepatology, 13(1):94-108, 2021.
- Porras P, Barrera E, Bridge A, Del-Toro N, Cesareni G, Duesbury M, Hermjakob H, Iannuccelli M, Jurisica I, Kotlyar M, Licata L, Lovering RC, Lynn DJ, Meldal B, Nanduri B, Paneerselvam K, Panni S, Pastrello C, Pellegrini M, Perfetto L, Rahimzadeh N, Ratan P, Ricard-Blum S, Salwinski L, Shirodkar G, Shrivastava A, Orchard S. Towards a unified open access dataset of molecular interactions. Nat Commun. 11(1):6144, 2020.
- Glaus Garzon JF, Pastrello C, Jurisica I, Hottiger MO, Wenger RH, Borsig L. Tumor cell endogenous HIF-1α activity induces aberrant angiogenesis and interacts with TRAF6 pathway required for colorectal cancer development. Neoplasia. 22(12):745-758, 2020.
- Yao Z, Aboualizadeh F, Akula I, Snider J, Tang P, Kotlyar M, Jurisica I, Stagljar I. Split Intein-Mediated Protein Ligation, a Novel Method for Detecting Protein-Protein Interactions and Their Inhibition, Nat Commun, 11(1):2440, 2020.
- Pinheiro M, Lupinacci FCS, Santiago KM, Drigo SA, Marchi FA, Fonseca-Alves CE, Andrade SCDS, Aagaard MM, Basso TR, Dos Reis MB, Villacis RAR, Roffa M, Hajj GNM, Jurisica I, Kowalski LP, Achatz MI, Rogatto SR. Cancers (Basel), 12(5):E1289, 2020. doi: 10.3390/cancers12051289.
- Reis PP, Tokar T, Goswami RS, Xuan Y, Sukhai M, Seneda AL, Moz LES, Perez-Ordonez B, Simpson C, Goldstein D, Brown D, Gilbert R, Gullane P, Irish J, Jurisica I, Kamel-Reid S. A 4-gene signature from histologically normal surgical margins predicts local recurrence in patients with oral carcinoma: clinical validation, Scientific Reports, 10(1):1713, 2020. doi: 10.1038/s41598-020-58688-y.
- Tokar, T., Pastrello, C., Jurisica, I. GSOAP: A tool for visualization of gene set over-representation analysis, Bioinformatics, Bioinformatics, 36(9):2923-2925, 2020. doi: 10.1093/bioinformatics/btaa001
- Rahmati R, Abovsky M, Pastrello C, Kotlyar M, Lu R, Cumbaa CA, Rahman P, Chandran V, Jurisica I, pathDIP 4: an extended pathway annotations and enrichment analysis resource for human, model organisms and domesticated species, Nucl Acids Res, 48(D1): D479–D488, 2020, https://doi.org/10.1093/nar/gkz989
- Kennedy, S., Jarboui, M-A, Srihari, S, Raso, C, Bryan, K, Dernayka, L, Charitou. T, Bernal-Llinares, M, Herrera-Montavez, C, Krstic, A, Matallanas, D, Kotlyar, M, Jurisica, I, Curak, J, Wong, V, Stagljar, I, LeBihan, T, Imrie, L, Pillai, P, Lynn, M, Fasterius, E, Szigyarto, C. A-K, Breen, J, Kiel, C, Serrano, L, Rauch, N, Rukhlenko, O, Kholodenko, B, Iglesias-Martinez, L, Ryan, C, Pilkington, R, Cammareri, P, Sansom, O, Shave, S, Auer, M, Horn, N, Klose, F, Ueffing, M, Boldt, K, Lynn, D, Kolch, W, Extensive Rewiring of the EGFR Network in Colorectal Cancer Cells Expressing Transforming Levels of KRASG13D, Nat Commun, 11(1):499, 2020. doi: 10.1038/s41467-019-14224-9
- Enfield, K.S.S., Marshall, E.A., Anderson, C., Ng, K.W., Rahmati, S, Xu, Z. Fuller, M., Milne, K., Lu, D., Shi, R., Rowbotham, D. A., Becker-Santos, D.D., Johnson, F.D., English, J.C., MacAulay, C.E., Lam, S., Lockwood, W.W., Chari, R., Karsan, A., Jurisica, I., Lam, W.L., Epithelial tumor suppressor ELF3 is a lineage-specific amplified oncogene in lung adenocarcinoma, Nat Commun, 10(1):5438, 2019. doi:10.1038/s41467-019-13295-y
- Monette A, Bergeron D, Ben Amor A, Meunier L, Caron C, Mes-Masson AM, Kchir N, Hamzaoui K, Jurisica I, Lapointe R. Immune-enrichment of non-small cell lung cancer baseline biopsies for multiplex profiling define prognostic immune checkpoint combinations for patient stratification, J Immunother Cancer, 7(1):86, 2019. doi: 10.1186/s40425-019-0544-x
- Monette A, Morou A, Al-Banna NA, Rousseau L, Lattouf JB, Rahmati S, Tokar T, Routy JP, Cailhier JF, Kaufmann DE, Jurisica I, Lapointe R. Failed immune responses across multiple pathologies share pan-tumor and circulating lymphocytic targets, J Clin Invest, 2019 Mar 26;130. pii: 125301. doi: 10.1172/JCI125301
- Kaufmann KB, Garcia-Prat L, Liu Q, Ng SWK, Takayanagi SI, Mitchell A, Wienholds E, van Galen P, Cumbaa CA, Tsay MJ, Pastrello C, Wagenblast E, Krivdova G, Minden MD, Lechman ER, Zandi S, Jurisica I, Wang JCY, Xie SZ, Dick JE. A stemness screen reveals C3orf54/INKA1 as a promoter of human leukemia stem cell latency, Blood, 133(20):2198-2211, 2019. doi: 10.1182/blood-2018-10-881441.
- Mandilaras, V, Garg, S, Cabanero, M, Tan, Q, Pastrello, C, Burnier, J, Karakasis, K, Wang, L, Dhani, NC, Butler, MO, Bedard, PL, Siu, LL, Clarke, B, Shaw, PA, Stockley, T, Jurisica, I, Oza, AM. TP53 mutations in high grade serous ovarian cancer and impact on clinical outcomes: a comparison of next generation sequencing and bioinformatics analyses. Int J Gyn Cancer, Jan 18. pii: ijgc-2018-000087. doi: 10.1136/ijgc-2018-000087.
- del Toro N, Duesbury M, Koch M, Perfetto L, Shrivastava A, Ochoa D, Wagih O, Piñero J, Kotlyar M, Pastrello C, Beltrao P, Furlong LI, Jurisica I, Hermjakob H, Orchard S, Porras P. Capturing variation impact on molecular interactions in the IMEx Consortium mutations data set." Nat Commun, 10(1): 10, 2019.
- Li L, Guturi KKN, Gautreau B, Patel PS, Saad A, Morii M, Mateo F, Palomero L, Barbour H, Gomez A, Ng D, Kotlyar M, Pastrello C, Jackson HW, Khokha R, Jurisica I, Affar EB, Raught B, Sanchez O, Alaoui-Jamali M, Pujana MA, Hakem A, Hakem R., Ubiquitin ligase RNF8 suppresses Notch signaling to regulate mammary development and tumorigenesis, J Clin Invest, 128(10):4525-4542, 2018. doi: 10.1172/JCI120401
- Kotlyar, M., Pastrello, C., Malik, Z., Jurisica, I., IID 2018 update: context-specific physical protein-protein interactions in human, model organisms and domesticated species. Nucleic acids research, 47(D1):D581-D589, 2019.
- Singh, M., Venugopal, C., Tokar, T., McFarlane, N., Subapanditha, M. K., Qazi, M., Bakhshinyan, D., Vora, P., Murty, N., Jurisica, I., Singh, S. K., Therapeutic targeting of the pre-metastatic stage in human brain metastasis, Cancer Res, 2018. ePub 2018/07/11. DOI: 10.1158/0008-5472.CAN-18-1022.
- Tokar T, Pastrello C, Ramnarine VR, Zhu CQ, Craddock KJ, Pikor L, Vucic EA, Vary S, Shepherd FA, Tsao MS, Lam WL, Jurisica I Differentially expressed microRNAs in lung adenocarcinoma invert effects of copy number aberrations of prognostic genes. Oncotarget. 9(10):9137-9155, 2018.
- Paulitti A, Corallo D, Andreuzzi E, Bizzotto D, Marastoni S, Pellicani R, Tarticchio G, Pastrello C, Jurisica I, Ligresti G, Bucciotti F, Doliana R, Colladel R, Braghetta P, Di Silvestre A, Bressan G, Colombatti A, Bonaldo P, Mongiat M. The ablation of the matricellular protein EMILIN2 causes defective vascularization due to impaired EGFR-dependent IL-8 production affecting tumor growth, Oncogene, 37(25): 3399-3414, 2018.
- Tokar, T., Pastrello, C., Rossos, A., Abovsky, M., Hauschild, A.C., Tsay, M., Lu, R., Jurisica. I. mirDIP 4.1 – Integrative database of human microRNA target predictions, Nucl Acids Res, D1(46): D360-D370, 2018.
- Kotlyar M., Pastrello, C., Rossos, A., Jurisica, I., Prediction of protein-protein interactions, Current Protocols in Bioinf, 60, 8.2.1–8.2.14., 2017.
- Singh, M., Venugopal, C., Tokar, T., Brown, K.B., McFarlane, N., Bakhshinyan, D., Vijayakumar, T., Manoranjan, B., Mahendram, S., Vora, P., Qazi, M., Dhillon, M., Tong, A., Durrer, K., Murty, N., Hallet, R., Hassell, J.A., Kaplan, D., Jurisica, I., Cutz, J-C., Moffat, J., Singh, D.K., RNAi screen identifies essential regulators of human brain metastasis initiating cells, Acta Neuropathologica, 134(6):923-940, 2017.
- Wong, S, W. H., Pastrello, C., Kotlyar, M., Faloutsos, C., Jurisica, I. Modeling tumor progression via the comparison of stage-specific graphs, Methods, 132(1): 34-41, 2018. Available online 3 July 2017.
- Pastrello C, Tsay M, McQuaid R, Abovsky M, Pasini E, Shirdel E, Angeli M, Tokar T, Jamnik J, Kotlyar M, Jurisicova A, Kotsopoulos J, El-Sohemy A, Jurisica I. Retraction: Circulating plant miRNAs can regulate human gene expression in vitro. Sci Rep. 7:46826, 2017.
- Wang, D, Pham, NA, Tong, JF, Sakashita, S, Allo, G, Kim, L, Yanagawa, N, Raghavan, V, Wei, YH, To, C, Trinh, QM, Starmans, MHW, Chan-Seng-Ye, MA, Chadwick, D, Li, L, Zhu, CQ, Liu, N, Li, M, Lee, S, Ignatchenko, V, Strumpf, D, Taylor, P, Moghal, N, Liu, G, Boutros, PC, Kislinger, T, Pintilie, M, Jurisica, I, Shepherd, FA, McPherson, J, Muthuswami, L, Moran, MF, Tsao, MS. Molecular heterogeneity of non-small cell lung carcinoma patient-derived xenografts closely reflect their primary tumors, Int J Cancer, 140(3): 662-673, 2017.
- Pinheiro, M., Drigo, S.A., Tonhosolo, R., Andrade, S.C.S., Marchi, F.A., Jurisica, I., Kowalski, L.P., Achatz, M.I., Rogatto, S.R., HABP2 p.G534E variant in patients with family history of thyroid and breast cancer, Oncotarget, 8(25):40896-40905, 2017.
- Citron, F., Armenia, J., Barzan, L., Franchin, G., Polesel, J., Talamini, R., Sulfaro, S., Croce, C.M., Klement, W., Pastrello, C., Jurisica, I., Vecchione, A., Belletti, B., Baldassarre, G., A microRNA signature identifies SP1 and TGFbeta pathways as potential mediators of local recurrences in head and neck squamous carcinomas, Clin Cancer Res, 23(14):3769-3780, 2017
- Sokolina K, Kittanakom S, Snider J, Kotlyar M, Maurice P, Gandía J, Benleulmi-Chaachoua A, Tadagaki K, Wong V, Malty RH, Deineko V, Aoki H, Amin S, Riley L, Yao Z, Morató X, Otasek D, Kobayashi H, Menendez J, Auerbach D, Angers S, Pržulj N, Bouvier M, Babu M, Ciruela F, Jockers R, Jurisica I, and Stagljar I. Systematic protein-protein interaction mapping for clinically-relevant human GPCRs, Mol Sys Biol, 13(3): 918, 2017.
- Yao Z, Darowski K, St-Denis N, Wong V, Offensperger F, Villedieu A, Amin S, Malty R, Aoki H, Guo H, Xu Y, Iorio C, Kotlyar M, Emili A, Jurisica I, Babu M, Neel B, Gingras AC, and Stagljar I, A global analysis of the protein phosphatase interactome, Mol Cell, 65(2):347-360, 2017.
- Petschnigg J, Kotlyar M, Blair L, Jurisica I, Stagljar I, and Ketteler R, Systematic identification of oncogenic EGFR interaction partners, J Mol Biol, 429(2): 280-294, 2017.
- Rahmati, S., Abovsky, M., Pastrello, C., Jurisica, I. pathDIP: An annotated resource for known and predicted human gene-pathway associations and pathway enrichment analysis. Nucl Acids Res, 45(D1): D419-D426, 2017.
- Chehade, R., R. Pettapiece-Phillips, Salmena, L., Kotlyar, M., Jurisica, I., Narod, S. A., Akbari, M. R., Kotsopoulos, J. Reduced BRCA1 transcript levels in freshly isolated blood leukocytes from BRCA1 mutation carriers is mutation specific, Breast Cancer Res, 18(1): 87, 2016.
- Cierna, Z., Mego, M., Jurisica, I., Machalekova, K., Chovanec, M., Miskovska, V., Svetlovska, D., Hainova, K., Kajo, K., Mardiak, J., Babal, P. Fibrillin-1 (FBN-1) a new marker of germ cell neoplasia in situ, BMC Cancer, 16: 597, 2016.
- Becker-Santos, D.D., Thu, K.L, English, J.C., Pikor, L.A., Chari, R., Lonergan, K.M., Martinez, V.D., Zhang, M., Vucic, E.A., Luk, M.T.Y., Carraro, A., Korbelik, J., Piga, D., Lhomme, N.M., Tsay, M.J., Yee, J., MacAulay, C.E., Lockwood, W.W., Robinson, W.P., Jurisica, I., Lam, W.L., Developmental transcription factor NFIB is a putative target of oncofetal miRNAs and is associated with tumour aggressiveness in lung adenocarcinoma, J Pathology, 240(2):161-72, 2016.
- Stojanova, A, Tu, W.B., Ponzielli, R., Kotlyar, M., Chan, P.K., Boutros, P.C., Khosravi, F., Jurisica, I., Raught, B., Penn, L.Z. MYC interaction with the tumor suppressive SWI/SNF complex member INI1 regulates transcription and cellular transformation, Cell Cycle, 15(13): 1693-705, 2016.
- Li, Y-H, Tavallaee, G., Tokar, T., Nakamura, A., Sundararajan, K., Weston, A., Sharma, A., Mahomed, N. N., Gandhi, R., Jurisica, I., Kapoor, M. Identification of synovial fluid microRNA signature in knee osteoarthritis: Differentiating early- and late-stage knee Osteoarthritis. Osteoarthritis and Cartilage, 24(9): 1577-86, 2016.
- Cinegaglia, N.C., Andrade, S.C.S., Tokar, T., Pinheiro, M., Severino, F. E., Oliveira, R. A., Hasimoto, E. N., Cataneo, D. C., Cataneo, A.J.M., Defaveri, J., Souza, C.P., Marques, M.M.C, Carvalho, R. F., Coutinho, L.L., Gross, J.L., Rogatto, S.R., Lam, W.L., Jurisica, I., Reis, P.P. Integrative transcriptome analysis identifies deregulated microRNA-transcription factor networks in lung, adenocarcinoma, Oncotarget, 7(20): 28920-34, 2016.
- Vargas, A., Angeli, M., Pastrello, C., McQuaid, R., Li, H., Jurisicova, A., Jurisica, I., Robust quantitative scratch assay, Bioinformatics, 32(9):1439-40, 2016.
- Snider, J., Kotlyar, M., Saraon, P., Yao, Z., Jurisica, I., Stagljar, I. Fundamentals of protein interaction network mapping, Mol Sys Biol, 11(12):848, 2015.
- Pettapiece-Phillips*, R., Kotlyar*, M., Chehade, R., Salmena, L., Narod, S. A., Akbari, M., Jurisica, I., Kotsopoulos, J. Uninterrupted sedentary behavior downregulates BRCA1 gene expression, Cancer Prevention Res, 9(1): 83-8, 2016.
- Kotlyar, M., Pastrello, C., Sheahan, N., Jurisica, I. Integrated Interactions Database: Tissue-specific view of the human and model organism interactomes, Nucleic Acids Res, 44(D1): D536-41, 2016.
- Navab R, Strumpf D, To C, Pasko E, Kim KS, Park CJ, Hai J, Liu J, Jonkman J, Barczyk M, Bandarchi B, Wang YH, Venkat K, Ibrahimov E, Pham NA, Ng C, Radulovich N, Zhu CQ, Pintilie M, Wang D, Lu A, Jurisica I, Walker GC, Gullberg D, Tsao MS. Integrin a11b1 regulates cancer stromal stiffness and promotes tumorigenecity in non-small cell lung cancer, Oncogene, 35(15):1899-908, 2016.
- Agostini M, Janssen KP, Kim LJ, D'Angelo E, Pizzini S, Zangrando A, Zanon C, Pastrello C, Maretto I, Digito M, Bedin C, Jurisica I, Rizzolio F, Giordano A, Bortoluzzi S, Nitti D, Pucciarelli S. An integrative approach for the identification of prognostic and predictive biomarkers in rectal cancer. Oncotarget, 6(32): 32561-74, 2015.
- Singh M, Garg N, Venugopal C, Hallett RM, Tokar T, McFarlane N, Arpin C, Page B, Haftchenary S, Todic A, Rosa DA, Lai P, Gómez-Biagi R, Ali AM, Lewis A, Geletu M, Mahendram S, Bakhshinyan D, Manoranjan B, Vora P, Qazi M, Murty NK, Hassell JA, Jurisica I, Gunning P, Singh SK. STAT3 pathway regulates lung-derived brain metastasis initiating cell capacity through miR-21 activation. Oncotarget, 6(29): 27461-77, 2015.
- Agostini M, Zangrando A, Pastrello C, D'Angelo E, Romano G, Giovannoni R, Giordan M, Maretto I, Bedin C, Zanon C, Digito M, Esposito G, Mescoli C, Lavitrano M, Rizzolio F, Jurisica I, Giordano A, Pucciarelli S, Nitti D. A functional biological network centered on XRCC3: a new possible marker of chemoradiotherapy resistance in rectal cancer patients, Cancer Biol Ther, 16(8):1160-71, 2015.
- Stewart, E.L., Mascaux, C., Pham, N-A, Sakashita, S., Sykes, J., Kim, L., Yanagawa, N., Allo, G., Ishizawa, K., Wang, D., Zhu, C.Q., Li, M., Ng, C., Liu, N., Pintilie, M., Martin, P., John, T., Jurisica, I., Leighl, N.B., Neel, B.G., Waddell, T.K., Shepherd, F.A., Liu, G., Tsao, M-S. Clinical Utility of Patient Derived Xenografts to Determine Biomarkers of Prognosis and Map Resistance Pathways in EGFR-Mutant Lung Adenocarcinoma, J Clin Oncol, 33(22):2472-80, 2015.
- Camargo, J. F., Resende, M., Zamel, R., Klement, W., Bhimji, A., Huibner, S., Kumar, D., Humar, A., Jurisica, I., Keshavjee, S., Kaul, R., Husain, S. Potential role of CC chemokine receptor 6 (CCR6) in prediction of late-onset CMV infection following solid organ transplant. Clin Transplant, 29(6):492-8, 2015.
- Fortney, K., Griesman, G., Kotlyar, M., Pastrello, C., Angeli, M., Tsao, M.S., Jurisica, I. Prioritizing therapeutics for lung cancer: An integrative meta-analysis of cancer gene signatures and chemogenomic data, PLoS Comp Biol, 11(3): e1004068, 2015.
- Starmans, M.H., Pintilie, M., Chan-Seng-Yue, M., Moon, N.C., Haider, S., Nguyen, F., Lau, S.K., Liu, N., Kasprzyk, A., Wouters, B.G., Der, S.D., Shepherd, F.A., Jurisica, I., Penn, L.Z., Tsao, M.S., Lambin, P., Boutros, P.C. Integrating RAS status into prognostic signatures for adenocarcinomas of the lung. Clin Cancer Res, 21(6): 1477-86, 2015.
- Tu, W.B., Helander, S., Pilstål, R., Hickman, K.A., Lourenco, C., Jurisica, I., Raught, B., Wallner, B., Sunnerhagen, M., Penn, L.Z. Myc and its interactors take shape. Biochim Biophys Acta, 1849(5): 469-483, 2015.
- Dingar, D., Kalkat, M., Chan, M. P-K, Bailey, S.D., Srikumar, T., Tu, W.B., Ponzielli, R., Kotlyar, M., Jurisica, I., Huang, A., Lupien, M., Penn, L.Z., Raught, B. BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors, J Proteomics, 118:95-111, 2015.
- Wong, S. W. H., Cercone, N., Jurisica, I. Comparative network analysis via differential graphlet communities, Special Issue of Proteomics dedicated to Signal Transduction, Proteomics, 15(2-3):608-17, 2015.
- Vucic, E. A., Thu, K. T., Pikor, L. A., Enfield, K. S. S., Yee, J., English, J. C., MacAulay, C. E., Lam, S., Jurisica, I., Lam, W. L. Smoking status impacts microRNA mediated prognosis and lung adenocarcinoma biology, BMC Cancer, 14: 778, 2014.
- Lalonde, E., Ishkanian, A. S., Sykes, J., Fraser, M., Ross-Adam, H., Erho, N., Dunning, M., Lamb, A.D., Moon, N.C., Zafarana, G., Warren, A.Y., Meng, A., Thoms, J., Grzadkowski, M.R., Berlin, A., Halim, S., Have, C.L., Ramnarine, V.R., Yao, C.Q., Malloff, C.A., Lam, L. L., Xie, H., Harding, N.J., Mak, D.Y.F., Chu1, K. C., Chong, L.C., Sendorek, D.H., P’ng, C., Collins, C.C., Squire, J.A., Jurisica, I., Cooper, C., Eeles, R., Pintilie, M., Pra, A.D., Davicioni, E., Lam, W. L., Milosevic, M., Neal, D.E., van der Kwast, T., Boutros, P.C., Bristow, R.G., Tumour genomic and microenvironmental heterogeneity for integrated prediction of 5-year biochemical recurrence of prostate cancer: a retrospective cohort study. Lancet Oncology. 15(13):1521-32, 2014.
- Berlin, A., Lalonde, E., Sykes, J., Zafarana, G., Chu, K.C., Ramnarine, V.R., Ishkanian, A., Sendorek, D.H.S., Pasic, I., Lam, W.L., Jurisica, I., van der Kwast, T., Milosevic, M., Boutros, P.C., Bristow, R.G.. NBN Gain Is Predictive for Adverse Outcome Following Image-Guided Radiotherapy for Localized Prostate Cancer, Oncotarget, 5(22): 11081–11090, 2014.
- Lapin, V., Shirdel, E., Wei, X., Mason, J., Jurisica, I., Mak, T.W., Kinome-wide screening of HER2+ breast cancer cells for molecules that mediate cell proliferation or sensitize cells to trastuzumab therapy, Oncogenesis, 3(12): e133; doi:10.1038/oncsis.2014.45, 2014.
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma, Nature, 511(7511):543-50, 2014. doi: 10.1038/nature13385. Epub 2014 Jul 9.
- Kotlyar M., Pastrello C., Pivetta, F., Lo Sardo A., Cumbaa, C., Li, H., Naranian, T., Niu Y., Ding Z., Vafaee F., Broackes-Carter F., Petschnigg, J., Mills, G.B., Jurisicova, A., Stagljar, I., Maestro, R., & Jurisica, I. In silico prediction of physical protein interactions and characterization of interactome orphans, Nat Methods, 12(1):79-84, 2015. E-pub 2014/11/18.
- Pastrello, C., Pasini, E., Kotlyar, M., Otasek, D., Wong, S., Sangrar, W., Rahmati, S., Jurisica, I., Integration, visualization and analysis of human interactome, J Biochemical & Biophysical Research Communications, 445(4):757-73, 2014. doi: 10.1016/j.bbrc.2014.01.151
- Cervigne, N.K., Machado, J. Goswami, R. S., Sadikovic, B., Bradley, G., Perez-Ordonez, B., Galloni, N.N., Gilbert, R., Gullane, P., Irish, J.C., Jurisica, I., Reis, P.P., Kamel-Reid, S. Recurrent genomic alterations in sequential progressive leukoplakia and oral cancer: drivers of oral tumorigenesis? Hum Mol Genet, 23(10):2618-28, 2014; doi: 10.1093/hmg/ddt657
- Petschnigg, J., Groisman, B., Kotlyar, M., Taipale, M., Zheng, Y., Kurat, C., Sayad, A., Sierra, J., Mattiazzi Usaj, M., Snider, J., Nachman, A., Krykbaeva, I., Tsao, M.S., Moffat, J., Pawson, T., Lindquist, S., Jurisica, I., Stagljar, I. Mammalian Membrane Two-Hybrid assay (MaMTH): a novel split-ubiquitin two-hybrid tool for functional investigation of signaling pathways in human cells; Nat Methods, 11(5):585-92, 2014; doi: 10.1038/nmeth.2895
- Der S.D., Sykes J., Pintilie M., Zhu C.Q., Strumpf D., Liu N., Jurisica I., Shepherd F.A., Tsao M.S. Validation of a histology-independent prognostic gene signature for early-stage, non-small-cell lung cancer including stage IA patients. J Thorac Oncol, 9(1):59-64, 2014. doi: 10.1097/JTO.0000000000000042
- Cirilo, P. D. R., Marchi, F. A., Filho, M. C. B., Rocha, R. M., Domingues, M. A. C., Jurisica, I., Pontes, A., Rogatto, S. R. An integrative genomic and transcriptomic analysis reveals potential targets associated with cell proliferation in uterine leiomyomas, PLoS One, (3):e57901, 2013.
- Vafaee, F., Rosu, D., Broackes-Carter, F. and Jurisica, I. Novel semantic similarity measure improves an integrative approach to predicting gene functional associations, BMC Sys Biol, 7:22, 2013.
- Goswami, R. S., Atenafu, E. G., Xuan, Y., Waldron, L., Pintor dos Reis, P., Sun, T., Datti, A., Xu, W., Kuruvilla, J., Good, D. J., Lai, R., Church, A. J., Lam, W., Baetz, T., LeBrun, D. P., Sehn, L. H., Farinha, P., Jurisica, I., Bailey, D. J., Gascoyne, R. D., Crump, M., and Kamel-Reid, S. A microRNA signature obtained from the comparison of aggressive to indolent non-Hodgkin lymphomas can be used in the prognosis of mantle cell lymphoma, J Clin Oncol, 31(23):2903-11, 2013.
- Berger T, Ueda T, Arpaia E, Chio II, Shirdel EA, Jurisica I, Hamada K, You-Ten A, Haight J, Wakeham A, Cheung CC, Mak TW. Flotillin-2 deficiency leads to reduced lung metastases in a mouse breast cancer model. Oncogene, 32(41):4989-94, 2013.
- Pastrello, C., Otasek, D.,Fortney, K.,Agapito, G., Cannataro, M., Shirdel, E.A., Jurisica, I. Visual data mining of biological networks: one size does not fit all, PLoS Comp Biol, 9(1): e1002833, 2013.
- Fortney, K., Xie, W.,Kotlyar, M., Griesman, J., Kotseruba, J., Jurisica, I. NetwoRx: Connecting drugs to networks and phenotypes in S. Cerevisiae, Nucl Acids Res, 41(D1): D720-7, 2013.
- Hammerman, P. S., et al., The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature, 489, 519–525, 2012.
- McKee, C.M., Xu, D., Cao, Y., Kabraji, S., Allen, D., Kearsmans, V., Beech, J., Smart, S., Hamdy, F., Ishkanian, A., Sykes, J., Pintile, M., Milosevic, M., Kwast, T. van der, Zafarana, G., Ramnarine, R., Jurisica, I., Mallof, C., Lam, W., Bristow, R.G., Muschel, R.J. Protease Nexin 1 modulates prostate adenocarcinoma by regulating the Hedgehog pathway. J Clin Invest, 122(11):4025-36, 2012.
- Kotlyar, M., Fortney, F. and Jurisica, I. Network-based characterization of drug-regulated genes, drug targets, and toxicity. Methods, 57(4): 477-485, 2012.
- Hai, J., Zhu, C. Q., Bandarchi-Chamkhaleh, B., Wang, Y. H., Navab, R., Shepherd, F. A., Jurisica, I., Tsao, M. S., L1 Cell Adhesion Molecule promotes tumorigenicity and metastatic potential in non-small-cell lung cancer, Clin Cancer Res, 18(7):1914-1924, 2012.
- Arneson, N., Moreno, Iakovlev, J.V., Ghazani, A., Warren. K., McCready, D., Jurisica, I. and Done, S.J. Comparison of whole genome amplification methods for analysis of DNA extracted from microdissected early breast lesions in formalin-fixed paraffin-embedded tissue, ISRN Oncology, v. 2012, Article ID 710692.
- Locke, J. A., Zafarana, G., Malloff, C.A., Lam, W. L., Sykes, J., Pintilie, M., Ramnarine, V.R.,Jurisica, I., Guns. E. T., van der Kwast, T., Milosevic, M., Bristow, R.G. Allelic loss of the loci containing StAR is prognostic for relapse in intermediate-risk prostate cancer, Prostate, 18(1): 308-316, 2012.
- Zafarana, G., Ishkanian, A.S., Malloff, C.A., Locke, J.A., Sykes, J., Thoms, J., Lam, W.L., Squire, J.A., Yoshimoto, M., Ramnarine V.R.,Jurisica, I., Milosevic, M., Pintilie, M., van der Kwast, T., Bristow, R.G. Copy number alterations of c-MYC and PTEN are prognostic factors for relapse following prostate cancer radiotherapy, Cancer, 118(16): 4053-4062, 2012.
- Locke, J.A., Zafarana, G., Ishkanian, A.S., Milosevic, M., Thoms, J., Have, C.L., Malloff, C.A., Lam, W.L., Squire, J.A., Pintilie, M., Sykes, J., Ramnarine, V.R., Meng, A., Ahmed, O., Jurisica, I., van der Kwast, T., Bristow, R.G. NKX3.1 haploinsufficiency is prognostic for prostate cancer relapse following image-guided radiotherapy, Clinical Cancer Research, 18(1): p. 308-16, 2012.
- Waldron, L., Pintilie, M., Tsao, M.S., Shepherd, F.A., Huttenhower, C., and Jurisica, I. Optimized application of penalized regression methods to diverse genomic data, Bioinformatics, 27(24): 3399-3406, 2011.
- Hakem, A., Bohgaki, M., Lemmers, B., Tai, E., Salmena, L., Matysiak-Zablocki, E., LópezBähr, W. I., Karaskova, J., Boutros, P., Sheng, Y., Arrowsmith, C., Chesi, M., Bergsagel, L., Perez-Ordonez, B., Squire, J., Jurisica, I., Penn, L., Sanchez, O., Benchimol, S., Hakem, R. Role of Pirh2 in mediating the regulation of p53 and c-Myc, PLoS Genetics, 7(11): e1002360, 2011.
- Reis, P.P., Waldron, L., Perez-Ordonez, B., Pintilie, M., Galloni, N., Xuan, Y., Cervigne, N.K., Warner, G.C., Makitie, A.A., Simpson, C., Goldstein, D., Brown, D., Gilbert, R., Gullane, P., Irish, J., Jurisica, I., and Kamel-Reid, S. A gene signature in histologically normal surgical margins is predictive of oral carcinoma recurrence. BMC Cancer, 11:437, 2011.
- Djebbari, A., Ali, M., Otasek, D., Kotlyar. M., Fortney, K., Wong, S., Hrvojic, A. and Jurisica, I. NAViGaTOR: Scalable and Interactive Navigation and Analysis of Large Graphs. Internet Mathematics, 7(4): 314-347, 2011.
- Eppert, K., Takenaka, K., Lechman, E.R., Waldron, L., Nilsson, B., van Galen, P., Metzeler, K., Poeppl, A., Ling, V., Beyene, J., Canty, A.J., Danska, J.S., Bohlander, S.K., Buske, C., Minden, M.D., Golub, T.R., Jurisica, I., Ebert, B.L., Dick, J.E. Stem cell gene expression programs influence clinical outcome in human leukemia, Nat Medicine, 17(9): 1086-1093, 2011.
- Notta, F., Doulatov, S., Laurenti, E., Poeppl, A., Jurisica, I. and Dick, J.E. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment, Science, 333, 218-221, 2011.
- Reis, P.P., Waldron, L.,Goswami, R.S., Xu, W., Xuan, Y., Ordonez, B.P., Patrick Gullane, P., Irish, J., Jurisica, I. and Kamel-Reid mRNA, S. mRNA transcript quantification in archival samples using multiplexed, color-coded probes, BMC Biotechnology, 11:46, 2011.
- Navab R., Strumpf D., Bandarchi B., Zhu C.Q., Pintilie M., Ramnarine V.R., Ibrahimov E., Radulovich N., Leung L., Barczyk M., Panchal, D., To, C., Yun, J. J., Der, S., Shepherd, F. A., Jurisica, I., Tsao, M. S. Prognostic gene-expression signature of carcinoma-associated fibroblasts in non-small cell lung cancer. PNAS, 108(17):7160-7165, 2011.
- Elschenbroich, S., V. Ignatchenko, B. Clarke, S.E. Kalloger, P.C. Boutros, A.O. Gramolini, P. Shaw, I. Jurisica, and T. Kislinger, In-depth proteomics of ovarian cancer ascites: combining shotgun proteomics and selected reaction monitoring mass spectrometry. J Proteome Res, 10(5): p. 2286-99, 2011.
- Fortney, K., and Jurisica, I. Integrative computational biology for cancer research. Human Genetics, 130(4): 465-481, 2011.
- Chang, Q., Jurisica, I., Do, T., Hedley, D.W. Hypoxia predicts for aggressive growth and spontaneous metastasis formation from orthotopically-grown primary xenografts of human pancreatic cancer, Cancer Res, 71(8):3110-3120, 2011.
- Shirdel, E.A., Xie, W., Mak, T.W., Jurisica, I. NAViGaTing the micronome: Using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS ONE, 6(2): e17429, 2011.
- Wei, Y.; Tong, J.; Taylor, P.; Strumpf, D.;Ignatchenko, V.; Pham, N. A.; Yanagawa, N.; Liu, G.; Jurisica, I.; Shepherd, F. A.; Tsao, M. S.; Kislinger, T.; Moran, M. F., Primary Tumor Xenografts of Human Lung Adeno and Squamous Cell Carcinoma Express Distinct Proteomic Signatures. J Proteome Res, 10(1):161-174, 2011.
- Jurisica I. Integrative Computational Biology, AI, and Radiomics: Building Explainable Models by Integration of Imaging, Omics, and Clinical Data, In: Patrick Veit-Haibach, Ken Herrmann (Eds), Artificial Intelligence/Machine Learning in Nuclear Medicine and Hybrid Imaging, Pages 171-189, Springer Cham, 2022. ISBN978-3-031-00118-5
- Kotlyar M, Wong SWH, Pastrello C, Jurisica I. Improving Analysis and Annotation of Microarray Data with Protein Interactions. Methods Mol Biol. 2022;2401:51-68. doi: 10.1007/978-1-0716-1839-4_5.
- Pastrello C, Niu Y, Jurisica I. Pathway Enrichment Analysis of Microarray Data. Methods Mol Biol. 2022;2401:147-159. doi: 10.1007/978-1-0716-1839-4_10.
- Pastrello C, Kotlyar M, Jurisica I., Informed Use of Protein-Protein Interaction Data: A Focus on the Integrated Interactions Database (IID). Methods Mol Biol., 2074:125-134, 2020. doi: 10.1007/978-1-4939-9873-9_10.
- Hauschild, A-C, Pastrello, C, Kotlyar, M and Jurisica, I. Protein-protein interaction data, their quality, and major public databases. Ed. N. Przulj. Analyzing Network Data in Biology and Medicine, An Interdisciplinary Textbook for Biological, Medical and Computational Scientists, Cambridge University Press, Cambridge, UK, pp.151-192, 2019. ISBN 978-1-108-43223-8. DOI: 10.1017/978110837770
- Wong, S., Pastrello, C., Kotlyar, M., Faloutsos, C., Jurisica, I. SDREGION: Fast spotting of changing communities in biological networks. Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 867-875, 2018.
- Kotlyar, M., Pastrello, C., Rossos, A., Jurisica, I. Protein-protein interaction databases. In: Ranganathan, S., Nakai, K., Schönbach C. and Gribskov, M. (eds.), Encyclopedia of Bioinformatics and Computational Biology, vol. 1, pp. 988–996. Oxford: Elsevier, 2018.
- Rahmati, S., Pastrello, C., Rossos, A., Jurisica, I. Two Decades of Biological Pathway Databases: Results and Challenges, In: Ranganathan, S., Nakai, K., Schönbach C. and Gribskov, M. (eds.), Encyclopedia of Bioinformatics and Computational Biology, vol. 1, pp. 1071–1084. Oxford: Elsevier, 2018.
- Hauschild, AC, Pastrello, C., Rossos, A., Jurisica, I. Visualization of Biomedical Networks, In: Ranganathan, S., Nakai, K., Schönbach C. and Gribskov, M. (eds.), Encyclopedia of Bioinformatics and Computational Biology, vol. 1, pp. 1016–1035. Oxford: Elsevier, 2018.
- Jeanquartier, F., Jean-Quartier, C., Kotlyar, M., Tokar, T., Hauschild, A.-C., Jurisica, I., Holzinger, A., Machine Learning for In Silico Modeling of Tumor Growth, Machine Learning for Health Informatics, State-of-the-Art and Future Challenges, LNAI 9605, Springer: 415-434, 2016.
- Larsen, S.J., Alkaersig, F.G., Ditzel, L.J., Jurisica, I., Alcaraz, N., Baumbach, J. A Simulated Annealing Algorithm for Maximum Common Edge Subgraph Detection in Biological Networks, GECCO, ACM Press, 341-348, 2016.
- Veillette, C. J. H. and I. Jurisica. Precision Medicine for Osteoarthritis. Osteoarthritis. Pathogenesis, Diagnosis, Available Treatments, Drug Safety, Regenerative and Precision Medicine. Eds. M. Kapoor and N. Mahomed, Springer: 257-270, 2015.
- Otasek, D., Pastrello, C., Holzinger, A., Jurisica, I., Visual data mining: Effective exploration of the biological universe; Eds. Holzinger, A., Jurisica, I., Interactive Knowledge Discovery and Data Mining: State-of-the-Art and Future Challenges in Biomedical Informatics, Volume 8401, LNCS, SOTA, Springer, 19-33, 2014.
- Koehler Leman, Julia, Pawel Szczerbiak, P. Douglas Renfrew et al. Sequence-structure-function relationships in the microbial protein universe. Nature Communications (2023). DOI: 10.1038/s41467-023-37896-w.
- Hung, Ling-Hong and Ram Samudrala. Rice protein models from the Nutritious Rice for the World Project. (2016). DOI: 10.1101/091975.
- Hung, Ling-Hong and Ram Samudrala. fast_protein_cluster: parallel and optimized clustering of large-scale protein modeling data. Bioinformatics (2014). DOI: 10.1093/bioinformatics/btu098.
- Hung, Ling-Hong and Ram Samudrala. Accelerated protein structure comparison using TM-score-GPU. Bioinformatics (2012). DOI: 10.1093/bioinformatics/bts345.
- Hung, Ling-Hong, Michal Guerquin and Ram Samudrala. GPU-Q-J, a fast method for calculating root mean square deviation (RMSD) after optimal superposition. BMC Research Notes (2011). DOI: 10.1186/1756-0500-4-97.
- Mottin, Melina, Bruna Katiele de Paula Sousa, Nathalya Cristina de Moraes Roso Mesquita et al. Discovery of New Zika Protease and Polymerase Inhibitors through the Open Science Collaboration Project OpenZika. Journal of Chemical Information and Modeling (2022). DOI: 10.1021/acs.jcim.2c00596.
- Silva, Suely, Jacqueline Farinha Shimizu, Débora Moraes de Oliveira et al. A diarylamine derived from anthranilic acid inhibits ZIKV replication. Scientific Reports (2019). DOI: 10.1038/s41598-019-54169-z.
- Hernandez, Helen W., Melinda Soeung, Kimberley M. Zorn et al. High Throughput and Computational Repurposing for Neglected Diseases. Pharmaceutical Research (2019). DOI: 10.1007/s11095-018-2558-3.
- Mottin, Melina, Joyce Villa Verde Bastos Borba, Cleber Camilo Melo-Filho et al. Computational drug discovery for the Zika virus. Brazilian Journal of Pharmaceutical Sciences (2018). DOI: 10.1590/s2175-97902018000001002.
- Mottin, Melina, Joyce V.V.B. Borba, Rodolpho C. Braga et al. The A–Z of Zika drug discovery. Drug Discovery Today (2018). DOI: 10.1016/j.drudis.2018.06.014.
- Mottin, Melina, Rodolpho C. Braga, Roosevelt A. da Silva, Joao H. Martins da Silva, Alexander L. Perryman, Sean Ekins and Carolina Horta Andrade. Molecular dynamics simulations of Zika virus NS3 helicase: Insights into RNA binding site activity. Biochemical and Biophysical Research Communications (2017). DOI: 10.1016/j.bbrc.2017.03.070.
- Ekins, Sean, Alexander L. Perryman and Carolina Horta Andrade. OpenZika: An IBM World Community Grid Project to Accelerate Zika Virus Drug Discovery. PLOS Neglected Tropical Diseases (2016). DOI: 10.1371/journal.pntd.0005023.
- Ekins, Sean, John Liebler, Bruno J. Neves, Warren G. Lewis, Megan Coffee, Rachelle Bienstock, Christopher Southan and Carolina H. Andrade. Illustrating and homology modeling the proteins of the Zika virus. F1000Research (2016). DOI: 10.12688/f1000research.8213.2.
- Lopez, Steven A., Benjamin Sanchez-Lengeling, Julio De Goes Soares and Alán Aspuru-Guzik. Design Principles and Top Non-Fullerene Acceptor Candidates for Organic Photovoltaics. Joule (2017). DOI: 10.1016/j.joule.2017.10.006.
- Pyzer-Knapp, Edward O., Changwon Suh, Rafael Gómez-Bombarelli, Jorge Aguilera-Iparraguirre and Alán Aspuru-Guzik. What Is High-Throughput Virtual Screening? A Perspective from Organic Materials Discovery. Annual Review of Materials Research (2015). DOI: 10.1146/annurev-matsci-070214-020823.
- Pyzer-Knapp, Edward O., Kewei Li and Alan Aspuru-Guzik. Learning from the Harvard Clean Energy Project: The Use of Neural Networks to Accelerate Materials Discovery. Advanced Functional Materials (2015). DOI: 10.1002/adfm.201501919.
- Hachmann, Johannes, Roberto Olivares-Amaya, Adrian Jinich et al. Lead candidates for high-performance organic photovoltaics from high-throughput quantum chemistry – the Harvard Clean Energy Project. Energy Environ. Sci. (2014). DOI: 10.1039/C3EE42756K.
- Olivares-Amaya, Roberto, Carlos Amador-Bedolla, Johannes Hachmann, Sule Atahan-Evrenk, Roel S. Sánchez-Carrera, Leslie Vogt and Alán Aspuru-Guzik. Accelerated computational discovery of high-performance materials for organic photovoltaics by means of cheminformatics. Energy & Environmental Science (2011). DOI: 10.1039/c1ee02056k.
- Hachmann, Johannes, Roberto Olivares-Amaya, Sule Atahan-Evrenk et al. The Harvard Clean Energy Project: Large-Scale Computational Screening and Design of Organic Photovoltaics on the World Community Grid. The Journal of Physical Chemistry Letters (2011). DOI: 10.1021/jz200866s.
- Sokolov, Anatoliy N., Sule Atahan-Evrenk, Rajib Mondal et al. From computational discovery to experimental characterization of a high hole mobility organic crystal. Nature Communications (2011). DOI: 10.1038/ncomms1451.
Computing for Clean Water
Discovering Dengue Drugs
Drug Search for Leishmaniasis
FightAIDS@Home
GO Fight Against Malaria
Genome Comparison
Help Conquer Cancer
Help Cure Muscular Dystrophy
Help Defeat Cancer
Help Fight Childhood Cancer
Help Stop TB
Human Proteome Folding
Mapping Cancer Markers
Microbiome Immunity Project
Nutritious Rice for the World
OpenZika
The Clean Energy Project
for supporting WCG work
and are greatly appreciated.
In the fall of 2004 IBM launched the World Community Grid (WCG). The WCG was funded and run by IBM as a philanthropic project until February 2022. Over the past year, the WCG has been transferred to the Krembil Research Institute, University Health Network (UHN), located in Toronto, Canada. The WCG is now managed by Dr. Igor Jurisica and his team at UHN.
We are exceptionally grateful to IBM for their many years of financial and operational support. WCG continues to support open-source and open-data research and helps reduce computational time to allow scientists to address the world’s most pressing questions at no cost to the researchers. As an academic group, we face a significant financial challenge in providing the same level of support to the global research community.
Across 19+ years of WCG, over 800,000 volunteers like you have provided their unused computing resources to support the work of diverse researchers who in turn have conducted ground-breaking medical and environment-related research that is changing the world. We thank everyone who has contributed the spare compute cycles of their devices at home, and hope that you will continue to support WCG as we expand the number of projects and grow the size of the grid.
WCG Budget in 2025
WCG requires back-end servers and storage to distribute the work and collect the results across individual projects. We need a dedicated staff including technical and communication personnel to maintain and develop the code, manage the infrastructure and run the projects, and engage with scientists and volunteers. We do not have the significant backing as IBM provided until 2021 for the operation of the Grid. We continue to search for long-term partners that could help supplement our research funding we use for covering the operating cost of WCG.
As of March 27, 2025: CAD 150,000 secured research funds; CAD 21,512 from donations
Blue bar indicates in kind contributions/donations from our academic and corporate partners;
Red bar represents donations received through the UHN Foundation and Princess Margaret Cancer Foundation;
Green bar shows allocation from our lab's research funds.
The initial sponsorship from IBM and Distributive enabled us to manage our infrastructure and operating costs in 2021 and 2022 by providing about 50% match to our research funding. However, since 2023, we run WCG with only our research funding. We need additional support more than ever. If you have some contacts, please, let us know. If you could spare additional resources to support us - there are multiple ways of donating to WCG. We continue to support the operation from research funds, but that alone is not sufficient.
As of March 27, 2025 we have received $21,512.15 in donations - thank you for your additional support. We have a long way to go - but is a great start of the year (we would prefer to provide a more up-to-date information - but unfortumately, we cannot even get the Foundation to provide us with more frequent updates). Through the support and donation from our academic and corporate partners, we have fully resolved our server, storage and server room costs for this year. We have committed half of our required operating cost from our research funding, leaving us with a deficit of CAD 128,488.To explore additional fundraising options - I have joined and participate in the Team Ian Ride. Supporting any of the riders, including Igor Jurisica would supplement the WCG-MCM budget to cover the deficit. Thank you in advance. Notably, these donations are related to the MCM cancer project - and thus are handled by the Princess Margaret Cancer Foundation.
Together, there is much more we can do. But my group cannot do it with only our research funding. As an academic resource, WCG faces significant financial and technical challenges in providing the necessary level of support to the global research community. WCG needs your help. If you are already contributing your computing resources, we thank you. If you haven’t yet, you can sign up here, and help discoveries that save lives and address global problems:
Also, if you can, please consider donating to WCG – any amount of assistance is appreciated. While the WCG is exceedingly efficient in terms of operational cost, we still need funding for software development, communications, and maintaining the infrastructure of the grid. If you, your friends and family, or your organization can help with a donation, this will go a long way to enabling WCG to continue and grow as the world’s largest volunteer-driven supercomputer, enabling seemingly impossible scientific research to come to life.
There are three options for donating to WCG:



Thank you for your support,
Igor Jurisica and the lab
- Upcoming Database Migration Will Require Taking BOINC Database Offline Between March 10th-11th - we have been testing the distributed Postgres architecture in our new staging environment. We intend to switch over to our Postgres build of the BOINC server v1.6.1 release next week, between March 10th-11th. The cutover will require a brief pause while we deploy the updated scheduler and feeder to each Postgres worker node, and rollout CDC via Kafka in production to match the staging deployment. With CDC, we will keep the legacy databases in sync with workunit state managed by the Postgres cluster.
- CRUD operations from create_work and validator_assimilator daemons will be pipelined by a journaling process to keep workunit state eventually consistent in the legacy BOINC database, so that the current API layer and business logic will continue to work as usual. What we gain is the IOPS available to the BOINC database will scale to the number of servers we deploy in the Postgres cluster, and batch updates to the database will preserve data locality, completing the horizontally scalable design.
- This should resolve the long-standing validation issues, high disk utilization for the current BOINC database, and enable us to return to strict homogenous redundancy classes for MCM1 workunits, solve the os_name and os_version buffer overflow and trunctation bug, and give us access to the new features implemented in the more recent BOINC releases, such as running containers as applications on the grid.
- We are able to produce and run MCM1 and MAM1 workunits through this system in staging, and will launch ARP1 shortly after we confirm it delivers as expected and described above. We also should then be able to release a larger quantity of workunits than we were previously able to for ARP1 and accelerate the completion of the project.
- Not a perfect state yet - but hopefully the systems are now stable enough. Congratulations to BOINC@AUSTRALIA for reaching 2,289,999 score on In Memory of Dylan Bucci - 2026 Challenge.
- there is still time to join Dylan's challenge - it runs till March 9: https://www.worldcommunitygrid.org/team/challenge/viewTeamChallenge.do?challengeId=11075
- We are rolling out BOINC server release 1.6.1 in production, as we successfully re-based our BOINC server build onto recently released upstream v1.6.1 and cherry-picked changes from legacy and new WCG development, passing all tests and adding new ones for WCG-specific customizations. The scheduler, transitioner, and feeder will now run alongside the create_work daemons, Redpanda brokers, file_upload_handlers, and combined validator_assimilator daemons on the BOINC backend cluster and serve only the node local partition, handling only BOINC client requests routed to that server by the load balancer.
- We are migrating the SQLite instances across the BOINC backend nodes to a Citus-Data v14.0 PostgreSQL database cluster, completing our transition to a horizontally scalable architecture for the BOINC backend via the Redpanda cluster, PostgreSQL database cluster, and our changes to enforce end-to-end data locality in workunit processing within individual partitions by changing signed URL generation to include buckets owned by a specific partition/node on the backend, and providing the load balancer a mapping of buckets to the correct backend node. While we have had bugs in multiple components as we made changes to the backend, all components now consistently apply the same hashing function to a given workunit ID to achieve correct routing, and as we loop the database and scheduler into this design we expect to finally be able to communicate the value of the new architecture in results as we restart ARP1, and finally rollout MAM1 and seek to backport the MAM1 application to the MCM1 project, for BOINC clients running on devices that can handle it. We will not close the path for less powerful devices to contribute, only add the path for more powerful devices to contribute more than they currently can.
- MAM1 v7.0.8 for Linux and Windows, and ARP1 will release as soon as the BOINC server v1.6.1 and distributed PostgreSQL database cluster rollouts are complete, we will then also enable stats export. We will be able to gather the remaining backlog for validation from the postgres cluster more easily than the currently bogged down legacy BOINC database, and perform the final phase of validation backlog reconciliation.
- Results page under My contribution, as correctly guessed in the forums, the botched rollout of the new results page was a direct attempt to reduce load on the BOINC database from the APIs now that result tables have grown so large, while attempting to preserve the ability of volunteers to inspect their results and confirm for themselves the validation state of their PV jails, which we have finally made progress on. After restoring basic functionality of the new server-side in-memory caching approach, we left several issues unresolved and proceeded to work on the above items. A fix is incoming to enable downloading all results instead of the 500 currently displayed, and to fix the conditions that cause the summary stats to show more results in the numerator than in the denominator, and to address the behaviour of filtering among others.
- Fixed Apache mod-security rules blocking Team Challenge and Invite to Challenge functionality, and we suspect multiple other 403 forbidden issues to be related to the upgrades to the Apache webserver and HAProxy load balancer at the start of the transition. More fixes are incoming for the website, once we have completed the rollout of the final pieces of our new horizontally scalable architecture, removing the BOINC database as a single point of contention, and launching all remaining applications as described above.
- Another crash in the data center - and more problems on WCG side.
- We are not yet sure if this is because Openstack at the data center has been running out of file descriptors for RabbitMQ or this is just the next "thought is was fixed" presentation of running out of file descriptors and we are headed for another crash
- We may be back up with a massive improvement soon. However, if it is the file descriptor issue, it may turn into another outage instead. Either way, Dylan is trying to use the downtime to improve the file_upload_handler, which will be the last component we still need the old BOINC build for. So, we'll be fully caught up on that side of technical debt or at least that is the plan.
- Another crash in the data center - this time - they hopefully identified the root cause.
- As of Feb 9 - web site up again; Dylan restarted the feeder, all six partitions back to active downloads and uploads; logs look good otherwise; restarted a stalled validator - should be all green now
- 11:02 PM ET: looks like data center outage again, from looks of it same as before when all our IPs dropped. Investigating.
- Outage Across All Web Services at worldcommunitygrid.org (We Think It Is DNS) - approximately 2:45 A.M. UTC our domain and all subdomains stopped resolving, although our servers and backend services remain operational. We do not yet know what the cause of the outage is, and are reaching out to the team that manages our DNS records for assistance.
- Phase 2/3 of MCM1 Validation Backlog Begins - last weekend into this week, we were able to successfully clear a few million more workunits from the MCM1 validation backlog without shutting down the BOINC database. Today, we have started the next phase, focusing on a different category of missed validations. As we run these validators on pre-compiled lists of result file pairs in parallel with the hot path (hot path uses the Kafka topics, firing result upload events so validator_assimilator daemons can batch up transactions of likely reported result pairs and try these "eligible" workunits efficiently), we have one more phase, Phase 3, planned for when Phase 2 completes. When Phase 2 and Phase 3 complete, that should "bring us current" to January 26th (most of the indexing was done on or before January 26th), and we can replay from the Kafka queues through the phase 1, phase 2, and phase 3 cleanup pipelines to get the rest from January 26th to the given day in February (should be a small fraction). The final "cleanup" phase will be re-evaluating workunits marked INVALID by each of the previous Phases 1-3, to see if recovery as VALID of some workunits with yet-more-relaxed criteria makes sense, especially given the large number of hr_class reset workunits we've been running to get around the host os_name and os_version issues from newer BOINC clients.
- We may still shutdown the BOINC database temporarily for MCM1 backlog validations - if we choose to do this to complete reconciliation, or to rollout the updated BOINC daemons based on the most recent server release 1.6.1, or go live with our distributed Postgres architecture to replace the current SQLite-per-partition architecture with a more robust, parallelizable backend, we will provide as much notice as possible. We will continue to reiterate this possibility while we are still considering it, as most services will not work if we take the database offline temporarily.
- BOINC db_dump for stats will resume once Phase 2 and Phase 3 backlog completes - we have not wanted to put out the stats, because we have not finished reconciling them from all the results that volunteers have uploaded, and we have not felt comfortable deleting results from the result table or result files from the caches or filesystems. We now know we can often credit retroactively, and once we are confident in our reconciliation of MCM1 backlog we will consider the pipeline and architecture stable, and resume db_dump stats dumps as before.
- We are pursuing upgrade to BOINC server release 1.6, which has the potential to fix the host os_version corruption issue (see https://github.com/BOINC/boinc/releases/tag/server_release%2F1.6%2F1.6.1) - we have seen entire error messages from the BOINC client end up in the os_name and os_version fields in addition to the most common case of a truncated single, first letter of the OS that should be recorded there. For example, "W" for Windows. While we cannot guarantee that this release will resolve the issue, and updating to it is still being assessed for feasibility and depends on other large upgrades, we are in a position to finally evaluate and attempt to switch off our legacy BOINC build to track upstream BOINC, and simply patch in the features required to maintain legacy continuity.
- MAM1, ARP1, MCM2 releases pushed back - the MCM1 validation backlog issues have stymied many of the cleanup processes required to keep adequate resources available to run multiple projects, especially with large upload sizes and local compute requirements like ARP1. We are in a position to release MAM1 workunits at any time. We have some scientific work to do before we begin distributing "MCM2" workunits using the same application MAM1 uses, as MCM1 batches. Once we are up to date with MCM1 backlog, and begin releasing stats, we will be ramping up other projects and begin approaching the frontend backlog.
- Issues with file_upload_handler on partitions 4, 5, and 6 resolves transient HTTP upload errors - this error generally reflects the file_upload_handler pool of Docker containers that Apache dispatches upload POST requests being in a bad state. As with the validators, assimilators, create_work daemons, file_deleter, db_purge - we will get the file_upload_handler out of a Docker container, after we fix the more pressing MCM1 validation backlog.
- MCM1 validation issue resolution may require BOINC database outage - we have concluded that freezing tables by taking the BOINC backend components and database offline to operate on a frozen copy of the key tables in the database would be the fastest and most surefire way to get the backlog validated. From each of the six servers we have referred to as "partitions" that own specific fanout buckets/non-overlapping ranges of workunits, we can assess the validation state of all uploads received by that partition independently against a copy of the result table, work out the exact upserts that will be required to idempotently set the correct state and if the workunit has not already been validated calculate and assign credit to volunteers, and then finally reconcile all six partitions patches and page the upsert against the "offline" (on the network) BOINC database to come back up in a fully resolved state.
- We have chenged the batch plan to have a shorter turnaround, most systems are easily computing within that time frame, and the notion of a "deadline" is significantly different now with the new validators where we greedily try to validate 2 results as soon as we have 2, but use the additional results if we have more than 2, and then assign everybody who is currently processing a result credit and call the workunit done.
- The point of the new deadlines is to get work done faster, so the two wingmen receiving results _0 and _1 get 3 days, then the transitioner might notice one or both haven't reported, and whoever downloads the resend _2 workunit gets 3 days to be the wingman, and so on every 3 days until we hit "maxError" (5), "maxCreatedForAnyReason" (10), "maxReturnedSuccessfully" (7). We don't want to keep sending workunits that are just errors no matter who gets them, if we've created it 10 times and someone has the _9 result something is wrong so stop, and if we have 7 results that we in theory could validate and credit already then something is wrong as we should be done after 2, which is what the new system emphasizes, or done after 3 assuming a wingman we thought was going to timeout actually pulls through sometime just after the deadline.
- A shorter deadline will help with resends finishing off workunits faster, especially once we are running MAM1 and MCM2 (and ARP1) that is going to make a big difference because instead of random searches, we are going to be doing targeted searches based on best results from the previous round. The faster we have results for the same set of parameters the faster we explore the search space. Resends will use the same delay bound as the workunit was created with, 3 days. The transitioner will refer to the record created in the database for the workunit, so indirectly it references the same parameter from the batch plan published to Redpanda.
- Regarding credit, we did not convey that part correctly. The IN_PROGRESS results and late uploads after deadline are not getting credit. The first pair above quorum (2), get credit, and so do any additional results that get uploaded and reported before the validator gets around to retrying the "eligible" workunit pool in chunks and happens to get to this set of results, ending the workunit and expiring any wingmen who were processing results that are not yet reported.
- So it is not "everybody who is currently processing a result gets credit", it is in fact "everybody who has already processed and reported a result, before the validator-assimilator daemon checked and found at least 2 SUCCESS results and effectively enforced the deadline".
- Outage at the data center has been resolved; we have regained access and completed recovery.
- Feeder has been restarted, and MCM1 workunits should resume distribution this evening.
- We were able to make some progress on the host info corruption bug, and establish a plan for a better workaround than the current reset of hr_class and floating point tolerant validation of the resulting resend cascade.
- We made some progress on the rollout plan to parallelize the feeder, scheduler, and BOINC database; this will happen soon. Benefit: stability and throughput of workunit distribution, especially once database is sharded across backend nodes and all workunit processing and state management becomes node-local. This is key for dealing with throughput of upcoming MDMG-family projects MAM1, MCM2 using LibTorch on GPUs.
- We worked on separating the "hot path" of newly created workunits downloaded, uploaded, and validated quickly on the same partition in-memory, versus the backfill events that require fetching results from other nodes and sometimes NFS to validate the pair. Although we deemed it too risky to deploy today we have made changes to parallelize across and within these two paths, per partition, to get back to rapid validation of newly created workunits while continuing the cleanup effort on the backlog which is currently too slow.
- We have lost access to the data center - trying to contact them - hopefully we will have some update soon.
- Fixed MCM1 credit calculation bug that was favoring the highest claimed credit; now correctly computing average for 2 results and median for 3+ results. Thank you to volunteers for reporting the discrepancy in the new validator_assimilator code for MCM1.
- Device profile updates from the website working again.A brief outage was required to restart the WAS server. Recent incidents appear to be related to MQ connection pool exhaustion or timeout issues. We will be able to focus on the growing list of stability issues with the website and forum once we have MAM1 running on Linux and Windows, and MCM1 validations caught up, which are the current focus of development. Apologies to users working on forum posts during the outage, we also have a candidate for the 403 error that will hopefully be resolved once we pivot to scheduled work on the website and forums.
- Warning: slow MCM1 validation as backlog validation continues. In addition to another round of architectural improvements to the distributed, partitioned validation and assimilation/accredation BOINC daemons, we have published to the ready for validation queue artifical events representing the location on the backend of validations uploaded to the wrong bucket due to the transitioner hashing resend upload URLs to the wrong server, HAProxy round-robin routing on redispatch and fall through on URL parse failure for initial uploads, and several additional edge cases that caused the pair of results computed and uploaded to be invisible to the new validation and assimilation process. Our fix adds fallback/fallthrough logic to the validator_assimilator daemon to facilitate remote file retrieval and process the tens of millions of backlog events we published to the queue it consumes from. We are exploring launching additional validator_assimilator daemons and separating backlog replay into dedicated Kafka topics to avoid slowing the hot path.
- Related to slow MCM1 validation, Redpanda data transforms that reduce upload events and emit pairs and resends for prospective validation went OOM during insertion of backlog events, requiring replay from the file_upload_handler topic that records single uploads as they hit the server. However, the replayed events reduced by the data transform are now AFTER the backlog events in the prospective vallidation queue. We should have spent the additional time and effort to create a separate path for backfilling the backlog of missing MCM1 validations, which would have avoided this unfortunate delay for those recently uploaded MCM1 workunits, but they will be credited.
- MAM1 smoke test batch MAM1_9999902 (LibTorch backend) validated successfully with ~80% success rate, every error was GLIBC < 2.29, not application bugs. Apologies for now cancelled batch MAM1_9999901, which was erroneously sent out using a defunct backend (dlib, for its SVM implementations) before we decided to focus on the LibTorch backend. Roughly 4.4% of active Linux x86_64 hosts are affected by the GLIBC requirement.
- Windows build of MDMG/MAM1 v7.08 successfully completed, deployment to beta30 in progress. We may have to include a large number of DLLs including pre-built LibTorch CPU libraries at first. These range in system requirements for Windows 10/11 64-bit with Visual C++ Redistributable 2015-2022, but for this initial build that successfully ran the test suite for the LibTorch backend in powershell on Windows 10, we expect that Visual C++ Redistributable 2022 is required. We've documented all third-party licenses (all permissive: BSD, MIT, Apache, NCSA, Zlib), and are hopeful a full static build will follow. System requirements are expected to be 4GB RAM or more, about ~300MB disk for application + DLLs, and we will be running the Windows build exclusively in beta for now starting sometime this week, and ramping up with the Linux build as we analyze first results.
- MAM1 beta testing forum topic and FAQ page will be up this week. We wanted to make sure we had the capability to create, send, receive, validate, cancel resends, and pick through results for both the beta30 project and production path before we committed to the launch process after many false starts in the past.
- Architectural Improvements to MCM1 and MDMG-family create_work and validator_assimilator daemons. We no longer build or run these daemons from a CentOS 7 base image Docker container, but directly on the Ubuntu 24.04 virtual machine hosts, and we have improved the in-memory cache of workunit metadata shared between the workunit creation and validation/assimilation process to use a dedicated SQLite instance, which also allows for live reaload of CLI parameters which are stored as metadata in a table such as the current offset into the ready for validation Kafka queue, and the delay between receiving a quorum of uploaded results and trying to assign credit to allow time for the scheduler to report outcomes to the BOINC database and avoid overly aggressive batch queries where many results have been uploaded, but not yet reported. We have also prepared for more nuanced validation of MDMG-family projects, where floating point tolerance can be adjusted based on aggregate statistics. A minimum count of identical gene signatures can be adjusted to deal with workunits where only some gene signatures were reported above the threshold value and similarly we can be lenient in validating workunits that had small total numbers of gene signatures "PASS" rendering aggressive validation critera moot if even acceptable floating point error causes the entire signature to be dropped. We have also facilitated "spike-in" canaries, allowing comparison of results for integrity of computation without strictly requiring the same parameterization, potentially "doubling" throughput on the overall problem space by allowing validation of results that mostly trained and evaluated models for non-overlapping seed gene signatures.
- SQLite instances for every running combination of mdmg_create_work and mdmg_validator_assimilator (--app is provided as CLI parameter) is a preliminary step towards distributed, sharded, partitioned postgreSQL, which is now a natural fit given our already partitioned workunit creation, routing of workunit downloads and uploads, and validation, which already guarantees non-overlapping batch commits crediting workunit results with the currently remote instance of MariaDB running the BOINC database. While there will always be potential contention in the user, host, and team table, and other sources, strict transactional boundaries ensure eventual consistency for these aggregations while result processing becomes scalable, and node-local once we make this move. Our intention is to align and time this final piece with upgrading the BOINC server backend to BOINC upstream, GPU builds for MCM1/MAM1/MDMG-family projects, and after fully testing in QA on a copy of the production data restored from backup to ensure that APIs can be pointed at the coordinator nodes of the new distributed postgres architecture without issue. We do not know exaclty when this will happen, but much of the ground work has been laid during the migration from Grahamcloud to Nibi, and we are looking to do this ASAP. We contend that without this kind of architectural overhaul, it will not be possible to keep up with the throughput of GPU-enabled MDMG-family projects, while adding new projects with collaborators.
- Pushed changes to BOINC transitioner to fix bug with upload bucket calculation for resends last night. This should drastically increase validations from resends going forward. We have also rebuilt the combined validation and assimilation pipeline for MCM1 and MAM1, which will finally enable us to start going through the validation backlog and clearing it out. We will try to leave a period of roughly 1-2 weeks for results to remain visible in a validated state (undeleted from the result table) as we validate and purge PV jail, which means removing the files from the in-memory cache and the result records from the database. We expect the process to take about 3 weeks.
- Working to resolve reported issue with profile changes again not propagating from website to BOINC clients the issue has a different presentation this time, we are working to resolve the issue.
- MAM1 beta workunits, and soon small smoke tests of the production pipeline, are being released - last week, we began smoke testing the MAM1 beta project (beta30) again, and we are working on the Windows and GPU-enabled builds in preparation for beginning the production run, keeping in mind the application we are using to run MAM1 will be backported to MCM1 as well so that we can take advantage of the modern features of the PyTorch/LibTorch backend. Thank you to volunteers for reporting outcomes in the forums across multiple threads. In the new year as we begin daily beta testing of MAM1 to roll out the initial production runs for the project and add platforms and GPU compat, we will have a dedicated thread for reporting issues, outcomes, and asking questions about the beta30 application and results on the forums.
- Thank You for supporting open science through WCG. Happy Holidays and Happy New Year to all volunteers!
- Forum service restored, after degraded service starting roughly 03:00 UTC, December 15th, 2025 led to a crash at roughly 12:30 UTC same day - service was restored at approximately 20:00 UTC Dec 15th, 2025. We have seen this before, due to database connections waiting indefinitely or so long as to eventually reach the thread pool maximum, causing OOM kill of the forum application under WAS, this is the meaning of the ForumUserServlet unable to initialize message displayed while the application was down. Previously, poor parameterization of the thread pool under WAS caused connections to the database to stay open instead of timing out under lock contention, we thought we had ameliorated the issue on the WAS side, but clearly this needs another look.
- At the very least, we will deploy alerts for the specific WARN and ERROR messages logged by WAS leading up to the crash which should provide a window of many hours within which we can fix this manually in the future, before the forum application crashes, pending a confirmed fix or workaround (e.g. better parameters and logic for managing thread pool).
- After announcing we would be accelerating validations "in the coming days", validations stalled again last week. Unfortunately, we continue to face issues with MCM1 validations. There are multiple categories of missed validations - orphaned "singles", mis-routed one or both results, incorrectly invalidated result pairs, missing resend condition, and now floating point tolerance too stringent for hr_class reset workunits which was the workaround to the impossible platform logjam issue, at the expense of having to validate workunits based on similarity of scientific results within a tolerance instead of strict equivalence of scientific results. The concept of validity for MCM1 became result pairs that have equivalent gene signature membership for signatures above the threshold score, and an equivalent list of gene signatures above the threshold score, and a similar score within a configurable error bound passed to the validator on startup, when the two workunits run with the same parameters and random seed value. While these cases should therefore be validated by the secondary validator we subscribed to the "validation failure" queue downstream of the primary, checksum based validator, our tolerance for floating point error was too stringent and we will be replaying the failure queue from an earlier offset to catch these cases for recently resent workunits.
- Regarding our approach to crediting workunits held during the downtime by scanning the filesystem and checking the database, the process began last week after indexing the locations of result files for all workunits across all filesystems on the backend, so that validations that involved file transfers could avoid the processing of walking remote filesystems and simply fetch the required remote result from wherever it had been uploaded or archived. Initial testing suggested we would catch most if not all missed validations using this approach, though the scripts each running on each worker node would have to run for some time. Clearly, despite thinking perhaps the timestamp-based approach was simply getting through points in time with few missing validations of any case early on, we are not making the expected progress. We are reviewing logs and stats on what has been processed so far to figure out what we missed, and how to adjust. Some validations have occured for each case, just nowhere near the expected throughput/hit rate we projected. So, we are tentatively hopeful we can fix this quickly and start finally making a dent.
- BOINC feeder/scheduler reporting "tasks committed to other platforms" is resolved - details are further down about the resolution and future plans to keep this issue from coming back.
- Validation Backlog has begun for workunits that were held over the break, and workunits that fell through our new validation logic unvalidated. We intend to ramp up these passes in the coming days, and will report on progress and project expected dates for fully backfilling all such cases and finally catching up validations to in flight work next week, now that we know our scripting works to backfill validations.
- We will not restart the file_deleter or db_purge BOINC services until we have validated every file we possess that was uploaded before/after the break, including sending resends for some cases of "orphans".
- What was the workaround for the feeder/scheduler blockage due to hr_class mismatch between results for the same workunit? The resolution to the issue that we chose for now, was to simply purge stale feeder entries effectively resetting their hr_class (homogenous redundancy) to 0 and allowing any host/platform to download the result if the result sits in memory for too long. The feeder can be started with a CLI option and specified time frame for occupancy of a result in a slot before it considers this course.
- What does resetting hr_class=0 as a workaround accomplish? The hr_class=0 reset matches the value assigned to fresh workunit results being sent out for the first time, essentially dictating to the scheduler that any host/platform may claim and compute this result (i.e., _0 and _1 results have hr_class=0, resends consult the hr_class of the host that reported results already). There is some computational overhead, as a second tier of validation is then required to validate the exact gene signatures and their scores are "the same" between these results computed on different platforms in the case of purged resends that had their hr_class reset to 0. We intend to disable hr_class (homogenous redundancy) completely for MCM1 at some point in the future, and instead rely directly on this currently secondary validation, and record of the delta between exact scores and verification of equivalent gene signatures found for these results sent to different platforms to ensure they are within a reasonable error bound/tolerance as a rule.
- Does this workaround affect the integrity of MCM1 results? No, but it does introduce a new edge cases to account for. The score can vary within the upper and lower bound of possible floating point error between platforms for the same workunit. Ensuring that the floating point calculations are not different enough to invalidate the computational result is a vastly easier problem when using the hr_class mechanism. However, because MCM1 produces a list of genes as well as a score, the only additional validation criteria we incur by disabling hr_class is ostensibly "score is just below the threshold on this system" exclusion, and "score is just above the threshold on this system" inclusion, for specific signatures very close to the configured threshold. In these cases, we can take the union of these additional results slightly above or below the threshold score, between all results for a workunit, provided the rest of the results above the threshold are equivalent.
- Why have hr_class at all for MCM1 then? Indeed. We intend to track the above cases and any other cases among validation failures where we can discern any unforseen effect of allowing resends to potentially go to different platforms, try this "disable hr_class if the feeder gets stuck" system for MAM1 which does have a numerical optimization routine to explore the signature search space that could change the actual signatures under test due to floating point error and so may not be a good candidate for this (and yet the calculations are valid, so any reasonable overlap or a "canary" or "spike-in" validation system might be considered sufficient validation...). If we are satisfied with the outcome of post-processing results that came from different platforms, we can disable it. This will accelerate throughput and discovery for MCM1 and possibly MAM1 while buying time to resolve this issue more permanently for applications such as ARP1 that this thinking does not apply to, where the floating point calculations must be byte-wise equivalent between results or the result is simply invalid. Once we can confirm that newer 8.x+ BOINC clients permitting WSL on Windows hosts is the only source of this hr_class confusion bug, and possibly the "W"/"W" os_name and os_version truncation bug, we can apply a targeted fix.
- We are testing required changes to the scheduler and feeder to resolve the corrupt/truncated "os_name" and "os_version" entries such as "W"/"W" for some hosts, as reported by users in the forums, and to resolve frequent "stuck" feeder states where "No tasks available for platform" is logically incorrect by hr_class, yet the tasks populating the feeder shared memory segment remain unassigned by the scheduler passes and manual intervention is required to get work flowing again.
- Passes through uploaded results that have not been credited by the new system will begin next week, to backfill missing credits. We have been performing dry runs to establish correctness. As a precaution, we will be running the program in multiple passes starting with the oldest uploads, to the most recent.
- Volunteers have reported that the API sometimes shows an invalid state for multiple results, where only one result is marked valid, which should be impossible. Preliminary investigation points to the new MCM1 assimilation procedure interacting with the transitioner. The new MCM1 assimilation procedure acts to validate and credit all in progress results for a workunit as soon as it has consumed any pair/quorum of files, whether original 0 and 1 results or resends 2 and up, that have passed validation. We will review this issue in full and report our findings, whether a bug in the assimilator, or poorly modeled interaction between assimilator transactions and the transitioner, which is where we expect to find an explanation.
- Database maintenance over Friday/Saturday completed without issue. We have resolved an issue with the backup scripts, effectively increased memory used to service database queries and added some new indices. We expect better performance from the BOINC database going forward.
- However, the disk remains slower than initial benchmarking when we stood up the database. We will monitor and reach out to hosting to see if the Ceph placement group expansion (that caused the stuck blocks of that particular disk when the placement group the result table lives on) got stuck in a "peering" state. We were informed that we should expect temporary, possibly intermittent slow IO during this Ceph maintenance window. If we can get faster disks for the BOINC database (which would require restoring the database to a new volume as we did to migrate) we will consider a maintenance window. Right now, we are optimistic the issues revealed in the new system by hanging database queries and database crashes can all be resolved with patches the new BOINC daemons, and current performance will be sufficient.
- As mentioned, this event identified several issues with the new BOINC daemons.
- MCM1 workunit creation proceeds in the Kafka topic even though the database is down, the mcm1_create_work daemon for it's Kafka partition on science01...science06 tries to commit it's part of the batch, database isn't there, so it doesn't do anything, but it does commit it's offset/pointer into the batch plan topic and move on to consume the next batch plan. That means every 10-15m while the database is down, a batch is effectively skipped. We were able to fix that, and have restarted MCM1 batch creation at roughly 5:00 p.m. EST, November 10th, 2025.
- We believe we have finally architected a fix for the pending validation backlog issue. This requires some non-trivial plumbing in the MCM1 batch assimilator, a Kafka connector deployed on the BOINC database node, and transitioner code changes.
- Workunit supply may remain artificially lower while we roll out the new batch assimilator builds and monitor the transitioner -> Kafka event consumption and result table interaction.
- We were able to resolve the issue with computing preferences not being updated from the website to BOINC client and vice versa. Generally, when the BOINC database goes down, so does the event listener that handles these messages on the webserver.
- We are still working on resolving the validation backlog from over the break, with the result table bricked during the Ceph maintenance we architected a "trust the filesystem" solution, and we are hopeful that this issue will be resolved this week.
- MAM1 was initially planned to be resumed in beta30 last week, to see if 7.07 fairly schedules work and respects --nthreads, which is a blocking issue in promoting the beta application to production. Depending on the error rate and behaviour on BOINC clients, we would then consider the stable code paths for the first production batches. Given our increased control over batch parameters with the new Kafka topic that uses a protobuf schema to fill out the workunit and result table entires, we intend to run work in production on Linux as soon as the beta30 application is stable with an error rate lower than MCM1 excepting the GLIBC dependency, which is typically the only repeated error we see from clients on the current LibTorch code path. We will then rely on iterating the beta30 application to 7.08 and 7.09 to get GPU and Windows support, and Parquet IO for input and uploaded results.
- We have fixed the main validation throughput issues with the new Kafka-based workflow, and reprocessed uploads from around the time we started sending out test batches. We are reviewing the Kafka topics and BOINC database to see if the volunteer reports of both results for a test workunit uploaded but no validation/assimilation occured during the reprocess is another bug to fix, and if so is it severe enough to block regular MCM1_024% batch distribution until resolved.
- In reviewing the transitioner implementation (which we intended to start yesterday to begin triggering resends for test batches), we found the new paradigm for storing configuration details that are required to populate resends in the result table needed to be incorporated into key functions. We are testing these relatively minor changes to the transitioner now.
- Our plan is to deploy the updated transitioner, verify resends work, verify it times out expired workunits, and depending on how that and the review of "missed validations" noted above goes we may then be ready to resume MCM1 batches in the normal range.
- Regarding uploads that span the downtime for migration, we will reconcile validation and credit for these workunits as soon as the production path for MCM1 described above is running. We should be able to use the new components to do that, after walking the filesystems where those uploads live, double-checking the list that need validation and crediting in the database, and pursuing a similar "reprocessing" path which worked well to re-attempt validation and crediting of the test MCM1 batches.
- Then, we will begin testing beta30/MAM1, and ARP1 using the new system, which we expect to progress much faster now that we have ironed out the logic with MCM1.
- tats updates will be restarted as soon as the MCM1 workflow is stable, that will include the daily export to https://download.worldcommunitygrid.org/boinc/stats/
- We have paused uploads and release of test batches while we work on the validation throughput issue.
- We think we have identified the root cause of the low validation rate for the MCM1 test batches, and we will send a few more test batches to confirm the fix does works.
- If this is the final fix and we see the expected validation rate for pairs of MCM1 uploads for new batches, we will replay the Kafka consumer on the upload events fired for test batches received earlier in the week, and this should idempotently allow the new batch assimilator to process those validations and assign credit.
- If the above goes well, we will schedule regular MCM1 batches to resume instead of the test batches.
- As volulnteers have noted, we have not yet reconciled uploads of regular MCM1 results submitted before we began sending test batches, and before the migration, but we have those files and will be able to do this in a batch update once the path for new workunits is working as described above.
- Naturally, we will resume ARP and MAM only after these issues are fully resolved.
- Finally stress testing rather than correctness testing.
- Sent a batch of 100,000 workunits (fast running, not full size in case something crashed.
- Thank you for your patience and continued support.
- We are sending small batches of workunits out starting tonight with batch IDs in the range 9999900+ for MCM1 to test the new distributed partition-aware batch upserting app-specific create_work daemons. The few volunteers who get these workunits before we start releasing larger batches as we gain confidence that the new system is working as expected may notice these workunits have a much smaller number of signatures and run much faster than normal. These are still meaningful workunits, but key parameters such as number of signatures to test per workunit were reduced so we could get feedback quckly.
- Similar to ARP1, we have moved all workunit templating and preparation to WCG servers for MCM1. We did this for the MAM1 beta (beta30) already, but we were able to move the rendering of workunit templates per batch into the create_work daemon C++ code directly, where it consumes a protobuf schema from Kafka/Redpanda's schema registry that it then hydrates to produce all workunits for the batch according to the desired parameters it consumes from the "plan" topic via Kafka. Hence, "app-specific" above. Then, it updates the BOINC database in bulk instead of calling BOINC's create_work() function. Metadata is local, partitioned, replicated in Kafka for durability, each batch writes files to that nodes' 1/6th of the buckets from the BOINC dir_hier fanout directory and commits 1/6th of the batch records to the database in non-overlapping ranges per 10k workunits per batch.
- The new validators are working and deployed. In our new distributed, partitioned approach, validators process workunits local to their host ONLY, uploads are partitioned according to the fanout directory assigned by BOINC, routed to the correct backend node by HAProxy corresponding to the BOINC fanout buckets. We split the buckets between nodes, instead of using them to fanout across the filesystem and avoid massive numbers of files in a single BOINC upload path, we fan out across the cluster and read/write these buckets in tmpfs so Apache serves downloads and accepts uploads in-memory, validators read in-memory, Kafka/Redpanda gets a copy of uploads into a disk-persisted, replicated topic for durability so if a node goes down and we lose the in-memory cache of downloads and uploads, we can replay and recover.
- By subscribing to a Kafka topic containing the count of uploads, a reduction on upload events emitted to Kafka topics from the new file_upload_handlers for only the local buckets of that partition, file locations pertaining to a pair of workunits, and emits success or failure to another queue for downstream "assimilation". We have written and are testing a batch applier that collects successful validation events on each partition, and batch updates the BOINC database so that the transitioner and scheduler can work together to evaluate the state of those workunits. Once we are confident the batch updates work as expected from the applier, users should start seeing workunits pending validation clear to valid.
- We are not running file_deleter or db_purge at the moment, they need to be rearchitected to match the new setup, or at minimum assessed to make sure it makes sense to start them unchanged. We have no concerns about running out of space in the database or on disk at the moment, only making mistakes, so we will get around to assessing what if anything needs to change about file_deleter and db_purge soon but not now. Likely, they will also take advantage of per-workunit event data from Redpanda/Kafka instead of just talking to the BOINC database and operate on local partitions across the cluster. But as we are producing events for every workunit's full lifecycle to Kafka topics we have a level of visibility and control we were never able to achieve with the legacy system, and we were able to set up prometheus node_exporter, tap into docker stats endpoints per node across the cluster, and likewise for Redpanda/Kafka with the helpful https://github.com/redpanda-data/observability repo to get a Grafana dashboard going that will let us do many things, such as serve up server status pages, and improve the stats pages.
- Testing the validators right now, been a lot of iterations on these.
- As soon as the validator works, we will deploy across the six partitions and clear the backlog. Then we can check the transitioner interaction. If that is all good, we can finally start sending new work.
- Going to finalize object storage for the archive - instead of previous tape backup.
- Happy Thanksgiving to our Canadian volunteers and partners.
- Work on finishing deployment setup will resume tomorrow.
- We have resolved the issue with the BOINC scheduler configuration causing "Another scheduler instance is running for this host". Users should be able to report tasks. We will update as soon as we begin creating new workunits as we are still working to stand up the rest of the BOINC backend architecture.
- Website went down briefly as we brought the scheduler online. We have adjusted the HAProxy configuration, and we will continue to adjust Apache/HAProxy config if we see the website stops responding again.
- Still debugging issues with the new Kafka-based validation workflow that works together with HAProxy routing rules to partition BOINC downloads and uploads by assigning servers equal hex buckets using the https://github.com/BOINC/boinc/wiki/DirHierarchy BOINC expects, and emitting events from the new file_upload_handler we wrote to Kafka so we can batch and respond to them in parallel. This removes the need for multiple round trips to the database for row-wise operations and polling, which are now simply batch applications of state after consuming workunits ready for validation in the relevant Kafka topic for that application. This allows us to perform validation and assimilation in the same process, at least for the projects we run ourselves (MCM1, MAM1, ARP1), and while the Kafka/Redpanda learning curve was significant, we have successfully transitioned to an event-driven in-memory partitioned architecture that should let us keep pace with the upcoming GPU enabled MAM1 application.
- We are aware of the issue with the scheduler returning "Another scheduler instance is running for this host" and have identified the cause in the config.xml template we adapated for the new containerzied environment. We will fix it once we have confirmed that the new event-driven validation and assmilation pipelines are working correctly.
- Uploads are being processed normally, we've confirmed the new architecture for the containerized file_upload_handler pool behind Apache is correctly producing to the per-application Kafka (Redpanda) topics, storing the event and result data in separate queues on the local brokers partition.
- As a result, there will be at least one more weekend sprint. Tentatively, we expect to be producing new workunits next week for MCM1, ARP1, and MAM1 beta version 7.07, validations should resume over the weekend, initial releases of batches will be intermittent.
- Hosting resolved the issue with our NFS shares in Openstack; scheduler and download path seem to work; we are debugging the new containerized file_upload_handler FCGI worker pool and file_upload_handler integration with Kafka through the librdkafka library.
- We will remove the ACL restrictions that are causing 403 forbidden when users try to contact the scheduler or upload files shortly.
- We are still validating the end-to-end upload path with our local BOINC clients. Once we confirm it, we will open up traffic to the upload servers, start serving the downloads that were available when we shutdown, and we can start turning the BOINC daemons back on.
- Tech team is working through issues with the WAS container configuration and the BOINC daemon builds on the science cluster.
- We are also pinging data centre admis as we don't have the /science filesystem on 1/6 of the science nodes, don't have the ability to add the IP address to the NFS share.
- Hopefully, not a blocker but does lock that node out of some roles until we can mount the NFS share.
- BOINC scheduler and uploads are projected to be up Monday, September 29th, 2025 after a final weekend sprint to test the new deployment strategy for increased performance, event driven horizontal scaling, and reduced load on the BOINC database.
- Forum issues detected.
- Sorry about these problems starting so soon after we came back - but - hopefully Dylan resolved them now.
- It all related to an AutoMaint features of DB2. Likely, database went into one of the scheduled maintenance tasks, MVNForum tried to talk to the database, didn't work, MQ blocked MVNForum and the lack of perms on the queue showed up as this init check failure.
- WCG migration is complete.
- Sorry about much longer downtime than anticipated - but - we are finally back online - and Dylan was able to make some improvements as well.
- We will share more details soon - but we also want to finish our start of the MAM project.
- Configuration of Websphere and IBM MQ is taking longer than expected. We are moving all provisioning, build, and deploy stages for all repos from Ansible and Gitlab CI to Dockerfiles and docker compose files, which is a step that precedes running these containers as StatefulSets on Kubernetes. So far, we have functional containers for IBM MQ, Websphere, DB2, MariaDB, and all BOINC endpoints up and running, and what we are still struggling through is configuration.
- This approach will benefit site reliability and scalability in an obvious way on Kubernetes, and will improve our development and QA lifecycles drastically. It was also necessary to preserve a maximum compatibility with the CentOS 7 virtual machines that the legacy stack was previously running on, a requirement for the redirected restore of the DB2 data for example, https://www.ibm.com/docs/en/db2/11.5.x?topic=restore-performing-redirected-operation.
- So why are we not up, and when will we be up? We are debugging the entrypoint scripts for Websphere and IBM MQ containers. Website cannot be brought up until Websphere is up and configured correctly, receiving messages from all MQ sidecars across the stack, sending emails, etc. Each of the databases, the webserver, and the scheduler have to run MQ, and we are still adapting some of the previous mqsc and other runtime configuration for the MQ service to work with this new setup where each important container that requires one gets an MQ sidecar container that uses the Ubuntu 24.04 host VM network.
- We are finalizing IBM MQ <-> DB2 <-> BOINC db <-> website axis, which will allow us to bring up the website. If all goes to plan now - we should have the website up tonight.
- Once that is solved - we will go through the BOINC stack to ensure nothing catastrophic will happen when once we let traffic through to the scheduler, upload/download servers. Then we can finally start letting the BOINC daemons manipulate state in the BOINC db.
- Over the weekend we were able to restore the DB2 databases for the website and forums.
- It was a redirected restore that first required a fully containerized instance of DB2 running the same OS as we were in Graham cloud, and we ran into issues attempting the restore of the final backups. Both databases are now successfully restored, and we have moved on to containerizing Websphere and IBM MQ.
- We were able to restore the BOINC database.
- As part of our work on MAM1 we developed an integration testing environment and containerized the BOINC database.
- We also did this for the BOINC server components (scheduler, upload and download servers with file_upload_handler, transitioner/validators/assimilators etc.).
- Once we get IBM MQ and Websphere up, we will be able to bring the entire system online shortly afterwards.
- Migration did run into some unexpected issues. No WU/results/credits will be lost. We will descrebe more, once we have some more time after the system is back on and running.
- Before we took the final backup of the BOINC database (Aug 31), all results had their deadlines extended by 96h, we expect to bring the BOINC database online today (Sep 4, 2025), and extend as needed until the system is operational
- Currently, WCG tech team is standing up DB2, IBM MQ, Websphere, and adpating Apache configuration.
- BOINC server components are expected to take less time to configure for operation at Nibi. We will stand up the upload/download server group ASAP so users can begin to upload results they have been holding on to during the migration, which will be credited.
- Full migration of WCG from the Graham to Nibi cloud facilities will be completed between 3:00-5:00 p.m. on August 31st, 2025
- Sharcnet will then power down all hardware at Graham.
- We have put in a ticket with UHN Digital to move our DNS records to the new IP addresses we have been allocated in Nibi cloud, and all storage, networking, and compute resources are already provisioned at Nibi.
- We continue testing QA and Prod on the new infrastructure.
- We will experience some downtime as *.worldcommunitygrid.org URLs switch over. We will be bringing down workunit creation scripting, BOINC server components, and upload/download servers in sequence, halting the database, performing a final rsync and then bringing down the website, forums, and internal services over the next 48h.
- In the best case, our DNS records will be switched over on the 31st and everything behind the load balancer will be up and running. However, we want to prepare users for the possibility of additional downtime as we stand up prod on Nibi.(
- MAM1 7.07 updates:
- The addition of spdlog as a dependency to replace the previous debug level printouts with more useful output for those who like to look at stderr.txt before it gets cleaned up by the BOINC client.
- Some kludgey math, flags, and options now set in the application's main function to try and get Ensmallen -> which depends on Armadillo -> which depends on OpenMP/OpenBLAS -> which nested thread creation causing suspension of concurrent running tasks under the BOINC client and using more CPU than the plan class and --nthreads parameter dictated. Essentially, bad behaviour. Thanks for posting feedback for the first few batches released, I will endeavour to address any further feedback if MDMG/MAM1 continues to over-schedule w.r.t the plan class or otherwise behaves badly.
- Added two built-in, configurable options for adjusting learning rate when using the LibTorch backend which was observed to improve the model's avg. loss progression during cross validation in fewer epochs, corresponding to:
- https://docs.pytorch.org/docs/stable/generated/torch.optim.lr_scheduler.ReduceLROnPlateau.html
- https://docs.pytorch.org/docs/stable/generated/torch.optim.lr_scheduler.CosineAnnealingLR.html
- Other fixes, additional work on features that will release in later versions after the migration.
- WU shortage resolved: WUs are coming back. Earlier today one of the disks filled up on the mesos cluster and that stopped the createWork daemon from running, it looks like ARP1 workunit input jobs were then forced to run on less of the nodes, backed up, preventing other jobs from scheduling on the nodes, those jobs failed, causing the process of downloading/building MCM1 work to ramp down to basically nothing since createWork couldn't run.
- MAM1 Beta 7.05 is in the final stages of alpha testing, debugging checkpointing issues under the BOINC client, upon release for linux will require GLIBC > 2.29: We forked the ensmallen library, "a high-quality C++ library for non-linear numerical optimization", and with cereal implemented serializable versions of Simulated Annealing (SA) and Particle Swarm Optimization (PSO) algorithms from the ensmallen library to supplement the existing random methods in MCM1 and MAM1.
- The new multi-stage MAM1 build compiles and statically links to LibTorch, OpenCV, dlib, and Apache Arrow for Parquet support, providing a state-of-the-art platform for general scientific inquiry and launching MAM1 and future projects. In addition to SVM, we now support RandomForest, AdaBoost, and Neural Networks (MLP/CNN) as classifiers to train and evaluate promising gene signatures identified by heuristic search of candidate gene signatures via SA and PSO.
- We have been working towards a CUDA-enabled LibTorch build for the application as well, and only linker errors remain before we can test setting torch::DeviceType to torch::kCUDA in place of torch::kCPU and see if it might be that easy.
- Kubernetes Migration of MCM1 WU Task Generation: We were able to confirm identical checksums and database updates for several batches between the existing and new approach to MCM1 WU delivery today. We are testing failure scenarios and planning the handoff of responsibilities from current infrastructure (remote UHN server generating tasks, WCG servers downloading, preparing/templating the input and XML files, tracking the tasks in the BOINC database as WUs and results). Likely, this will take a few days to complete.
- Testing: What remains to be done is some minor "chaos testing" on QA servers (killing the process, killing the box, DB drops the connection, etc), which we intend to subject the new all-in-one service that is replacing the download, template input files/build, and create work "commit new WU records to BOINC in batch" stages of the MCM1 workunit batch generation, and delivery pipeline.
- This is building on work we have already completed, which generates the MAM1 beta WUs, almost identical to the MCM1 WUs, directly on WCG servers. ARP1 also has all task inputs generated locally, and once this MCM1 migration is completed, we intend to move ARP1 job scheduling to the Kubernetes cluster as well, in addition to MAM1 and all future projects.
- MCM1 Workunit Availability: The recurring problem where older servers/VMs in our private cloud lose their DHCP lease on all interfaces and effectively go down, has been causing the quorum of coordinator nodes that accept jobs and assign them to workers ("the scheduler"), to stop accepting job submissions when this crash coincides with a second issue that renders the coordinators unable to recover their quorum. Building on the work we did to generate ARP1 and MAM1 workunits locally on WCG servers, we are migrating the MCM1 workunit delivery to Kubernetes, which should permanently resolve the issue, and increase workunit supply overall. Initially we had planned to complete this work after the release of MAM1 7.05, which has been a significant refactor, but given the frequency of failures we are moving it up and will complete the migration this week.
- MAM1 7.05 - Why is it Taking so Long? MAM1 is being refactored to use LibTorch and run also on NVIDIA GPUs. LibTorch vastly simplifies the checkpointing logic, and should resolve the unsafe memory access and resume from/checkpointing crashes in previous beta releases as well as significantly improve performance.
- 12:40 ET - WCG database crashed. Dylan is rebooting VM and hopefully we can recover without Sharcnet sys admins.
- Server is up. Assuming DB crash recovery goes well, we should be back online in 1-2 hours.
- BOINC database crash recovery was successful; service has been restored: DB recovered and restarted, all BOINC daemons and feeder have been restarted; reset the state of the scheduler coordinator and confirmed job submissions are working, verified download of new MCM1 and ARP1 workunits.
- MAM1 Beta Batches w/ Unacceptable Error Rate and Runtime: A small 4-work units batch (MAM1_9800035) to confirm the fix and a larger 100 work units batch (MAM1_9800036) were released yesterday night and this morning, respectively. So far no errors have been returned from either batch.
- Reported Missing Beta Results; Lack of Results on Community Stats Page: Between 2025-04-24 and 2025-04-25, several beta batches were released for Linux/Windows MAM1 application version 7.04, numbered between 9800000-9800026. The intent was to vary parameters to optimize the quality of signatures returned based on previous runs of MCM1 and our local testing. Unfortunately, these batches revealed multiple issues and in some cases exploded the runtime as many volunteers have reported in the forums. We have updated the workunit records in the BOINC database on 2025-04-25 for all these workunits in this range of batches to hopefully prevent resends and Server Abort any BETA workunits that were awaiting a free slot to begin execution on BOINC clients. For those who continue to run these long running workunits, we will monitor those that remain outstanding and continue to extend deadlines.
- New MAM1 Beta Batches 9800027-9800031 Released based on Low/No Error Rate Batches (9800001, 980008): We plotted outcomes for the 100-1000 workunit batches between 9800000-9800026, and released a series of batches 2025-04-25 from 9800027 onward that are based on those with no or low error rate. For these we have varied the length of the signature, and number of iterations, but left the model parameters known to be stable untouched. We will continue tuning the MAM1 model settings for the new dataset based on these preliminary distributions of signatures being returned by beta testers, and strictly confirm resource requirements and runtime of any changes to model parameters in the workunit settings file in all future MAM1 beta batches.
- Fixing OOM after suspending or power-cycling the machine running the BOINC client: We will fix these issues in version 7.05 of the MAM1 application next week.
- MAM1 Windows 64-bit build released in application version 7.04: We were able to resolve the memory safety issue around checkpointing and shared memory for the graphics application, and released a new version of MAM1 with the Windows x86_64 build. It is currently running on several Windows 10, Windows 11, and one Windows Server 2022 client. We will start sending larger batches that process more signatures, and use more stringent cross validation once the initial batch sent out to confirm the fix comes back with no or low error rate.
- Beta Batch MAM1_9800037 Released to Test Fix to Windows 64-bit Build: Another small batch of 100 workunits has been released with MAM1 7.04 based on the previous batches MAM1_9800035 and MAM1_9800036, which all returned without an error (208 workunits with some variation in code paths, number of signatures etc. amont the higher numbers).
- Graphics Images Display Issues in version 7.02-7.04: Users may notice that the TGA files that form the background of the composite image on which the chromosome map is rendered, and also the container for the chromosome map, are rendered differently between Linux and Windows and they are out of position. We will be iterating on the graphics as we progress through the beta, but our focus is on working builds for a maximum number of architectures and optimizing and stabilizing the singature permuation, SVM training, and signature evaluation code.
- ARP1 Stuck Extremes, low throughput: Now that we have Windows and Linux builds for MAM1, we will be able to spend some time trying to reproduce and resolve the ARP1 extreme generations that are no longer progressing, and start increasing throughput for ARP1 in general.
- MAM1 Beta Linux Build Fixed in version 7.02: We have fixed the issue causing crashes durining initialization with the Linux 64-bit build, and realeased an updated MAM1 application version 7.02 for Linux.
- MAM1 Beta Batches MAM1_9800035 and MAM1_9800036: A small 4-work units batch (MAM1_9800035) to confirm the fix and a larger 100 work units batch (MAM1_9800036) were released yesterday night and this morning, respectively. So far no errors have been returned from either batch.
- Reported Missing Beta Results, Lack of Results on Community Stats Page: As some volunteers have noted, we have not yet run the validators and assimilators for MAM1. We did write and build the validators and assimilators, which are essentially the same as those for MCM1. If there are no surprises, we expect to schedule these as daemons and let them run indefinitely assuming the initial batches of results validate without issue.
- Windows 64-bit Build Next: Primary effort at the moment is on resolving the previously mentioned issues with the Windows build of the MAM1 application. We plan to make a more formal post in the Beta Testing thread for reporting issues once we release the initial Windows version.
- MAM1 Beta Begins: We have (finally) started MAM1 beta testing on April 4th, 2025. Linux build first.
- Version 7.00 and 7.01 Crashes: First and second batches MAM1_9800033 and MAM1_9800034 of 100 work units each sent to 64 bit Linux clients crashed immediately with a segfault. Initially we thought this was a "regression" from pulling the wrong branch into the Docker container that we used to build MAM1 and graphics for Linux. However, version 7.01 intended to resolve the crash, which we had seen before in alpha testing; yet it did not. We will be pushing 7.02 to resolve these issues soon, and start releasing more batches then. Apologies for the oversight to all volunteers who ran with version 7.00 or 7.01 across these two batches.
- Windows Build Status: We were able to produce a working build for Windows 8.1 Pro to integrate with the Visual Studio 2008 and Visual Studio 2005 project solutions for Graphics and BOINC dependencies. However, testing on Windows 10 revealed memory safety issues around checkpointing and graphics. We are working to fix these issues before we push the first Windows 64-bit build of the MAM1 application.
- MacOS Build Status: Given a stable Linux build, we intend to finalize Intel-MacOS soon; we will work towards aarch64, amd64, arm64 once we have the Intel Macs covered.
- Optimizations: Now that we have been able to build and ship the application based on the MCM1 code and have the ability to iterate on the build and release new versions quickly, we will begin our efforts to optimize and modernize the code.
- We have successfully updated all WCG core libraries, BOINC, Boost, MCM1, and MAM1 in Visual Studio 2022 (formerly 2011/2013) to produce Windows builds for MCM1 and MAM1, prepare for BOINC server upgrade to track upstream BOINC, and to enable future application development for Windows clients. Previously, the toolset was v120, and the Visual Studio install either 2011 or 2013.
- Mac builds for MAM1 will be delayed slightly; we are exploring using Github Actions, CircleCI, and old unused Mac PCs from team members that we can install XCode on. The lab does not use Mac and we have limited hardware and experience available.
- We have began to work on reimplementing the createWork daemon and eliminate deadlock bug in multi-threaded preparation of MCM1 workunits set to be made available, when complete intermittent pauses in MCM1 availability should cease.
- createWork daemon problem - batches are not loading. We are working on fixing the issue.
- Interruption to workunit availability today for MCM1 and ARP1 should now be resolved.
- Issue was caused by a bug in the multi-threaded createWork process, under certain conditions deadlock results in new batches of workunits set to be marked available to be rolled back.
- We expect to resolve this issue permanently with the expanded capacity upcoming with the migration to Kubernetes.
- The DHCP lease issue, or whatever the root cause of our production VMs losing all network access at an increasing rate such that we are almost sure to experience a server crash multiple times a week, is being investigated by hosting.
- Our plan to resolve this regardless of the outcome of the investigation is to fully migrate most production boxes to Kubernetes including the DB2, Websphere, and IBM MQ "axis" of the website/forums and webservices provided by WCG.
- Previously, we had only provisioned QA on the Kubernetes cluster, and intended to further provision and deploy containers running Mesos workers as our first production boxes orchestrated by Kubernetes on the new hardware to blue/green deploy and eventually move the coordinator responsibilities and finally all workunit management pipeline responsibilities to Kubernetes running Mesos, which would give us fault tolerance at last as we pick apart all the old Mesos job descriptions and crontabs to fully migrate to Kubernetes, Slurm, Redpanda, and distributed postgres (Citus-Data).
- Once finished, we will decomission Aurora/Mesos and the old CentOS 7 boxes that run and coordinate the Mesos cluster, and provision new VMs with an LTS version of Ubuntu as we have on the new hardware to add that capacity to the Kubernetes cluster.
- We apologize for the delays to the start of the MAM project, we did not account for the sword of damocles hanging over every production server and falling with increasing frequency, nor for the reduced capacity of the environment in the new year. Thank you for your patience and understanding, we will be starting MAM shortly, as soon as we are through this issue.
- 4:27pm ET: db02 is back online; the website is back up; the DHCP agents were flushed on the old WCG nodes, which should resolve the issue
- Services seem to be down. We are working on identifying and fixing the issue.
- BOINC db node crashed. Thus, all running BOINC services, API services and message queues that need to talk to db01 die similarly; the connection is closed, although the node itself is still running.
- 10:38 am ET: Crash recovery starting now. We should be able to restart all the services soon.
- 12:21 pm ET: crash recovery successful; bounced all services; restarted the feeder; should start to see work going out again.
- Murphy's laws - one of the old servers passed away - and that was the "leader" - so work was available, but all systems were waiting for "the chosen one" to respond. While "soon" we should be moving to the new setup - we do hope it will be "soon enough" before another one dies.
- We have resolved the overnight outage of our job scheduler Aurora today, March 1st, 2025.
- Aurora is responsible for accepting requests to build new batches for MCM1, create new single ARP1 workunits, and running the BOINC component services in Mesos containers.
- Aurora stopped accepting job submissions due to a failure in one of the aurora-scheduler services that form the cluster "leader" quorum under Zookeeper.
- While batches had been building on schedule, a new leader was not automatically elected, and the cluster essentially went down stopping all new jobs from launching.
- We have confirmed that manual and automated builds via job submission of new MCM1 batches are going ahead, and will monitor the rest of the pipeline until we see workunits going out to users this afternoon.
- Multiple settings in the Aurora scheduler sysconfig have been identified that could result in this problem again. We will fix those.
- We expect to resolve this issue permanently by having Kubernetes manage minimum quorum until we can completely migrate off of Aurora and Mesos in favor of Kubernetes and Slurm. This will be implemented with our upcoming migration to Kubernetes running on new hardware to blue/green deploy our new tech stack.
- We are aware of the issue with sporadic generation of workunits. The cause is our job scheduling infrastructure, we are attempting to resolve this today and will update volunteers tomorrow on the result.
- Several jobs that we schedule on the backend write lockfiles that will prevent the job from running again, if the previous run is still ongoing. However, there are several points in the scripts that launch these jobs where a non-critical failure will result in the lockfile preventing future work from being indexed and created. We began experiencing these failures after we had multiple server "crashes" in the past two weeks, a lingering issue with the DHCP agent becoming "stuck", eventually resulting in non-responsive VMs as their DHCP leases expire and are not renewed. This has happened to multiple of the six workunit management servers where backend jobs to generate workunits are run, and one of the upload/download servers, in the past week or so. This has resulted in several problems, that we are trying to resolve to bring the system back into a stable state.
- The "modulo 4" issue identified with the MCM1 validators for example, was caused by one of these crashes. As surmised in the forums, the number of validators is scaled up by re-launching them as services on the Mesos cluster with a specific modulus. We have investigated the logs for the validators and despite the other issues we are facing with the Mesos cluster at the moment, we are seeing validations go through. The rate of validation should improve as we restore stability to the Mesos cluster and the services the jobs that run on Mesos interact with.
- Unfortunately, these problems are caused by part of the system running in the oldest servers (which we started to use in 2021), which we will move out of once the new system becomes available.
- We were able to compile and test a standalone build of the MAM project, incorporating the small group of WCG libraries that interface with the BOINC client API to handle IO, checkpointing, and graphics in the same way other WCG applications such as MCM1 do.
- The MAM1 application is essentially the MCM1 code with some improvements (and new graphics). We were able to generalize the data processing steps required to produce the custom dataset format that MCM1 currently supports. We will now be able to evaluate arbitrary normalized expression datasets from Gene Expression Omnibus (GEO) repository, and launch new projects searching for biomarkers much easier.
- Currently, we are working to deploy the MAM1 application in our QA environment to ensure that integration with BOINC, application graphics, and our scripts for generating and processing workunits locally will work as expected.
- There remain some issues with QA environment since construction at the data centre was completed. Although we were able to bring up production system on January 10, 2025, many of our QA servers were bricked. We are working with staff at Sharcnet to fix these problems, so that we can properly test the application. Once we are able to evaluate it in QA environment, we can export a satisfactory version from the QA BOINC database, and import into the production environment and get the beta started. We will keep you posted.
- All our servers are back online. We are downloding processed work units and sending out new ones, and they have the right deadlines (after the initial glitch with the time).
- We have noticed 92,194 results that ended up in an error state. We saved all these in a file so we can repair this issue. We'll be able to rescue these, especially since there is no file deleter or db purge daemon running for now, so all data related to these workunits is still on the filesystem.
- We will continue to monitor the system to make sure it is stable.
- We are also working on getting MAM project into beta.
- Thank you all for continuing to support research. Happy new year.
- BOINC database is up and in a good state. We are waiting on two more servers to regain access to the network, at which point we will be restarting the scheduler, transitioner, assimilators and validators.
- All deadlines for outstanding MCM1 work units have been extended to just after 6:00 p.m. Eastern Standard Time on January 15th, 2025.
- Web site is up; stats will be updated soon.
- Forums are up.
- Most of our infrastructure is back online. Unfortunately, some issues with the network and specific virtual machines remain. Thus, the BOINC database node remains unavailable, and the website and forums also do not function properly.
- Sharcnet data center team is working on to restore access to these instances in priority order.
- Once this is resolved, we will have a smooth restart of the workunit management and BOINC components on the backend, and be able to isolate and diagnose any remaining issues as we restart.
- Networking issues have been resolved. The data centre staff is finalizing our access.
- Update from the data centre: "Having issues with the physical network, likely can't get it diagnosed and fixed until Monday, January 6.". As a result - we still do not have a connection to our servers.
- We have been notified that the core system at SHARCNET is coming online now (5pm). They are planning to complete it tonight (January 3). We are waiting for the access to our systems, and will start turning everything back on as soon as we gain login.
- We have begun testing the Mapping Arthritis Markers (MAM1) prototype based on the current MCM1 code. We expect to launch the beta version of the application shortly after we are back online on the 3rd of January. We will provide a firm launch date once we are back online. The GPU version of the project may take some additional time to develop and test.
- We have been reviewing the MCM1 application code and considering alternative upgrade paths for MCM1 and related projects to enable GPU compute for gene signature search in general. We have decided to work towards replacing the SvmLightLib dependency and other related sections of the code with more modern, but still self-contained dependencies that will provide the opportunity to select CPU vs. GPU backend depending on user preferences and device capability. This should also enable us to take advantage of modern instruction sets and possibly other architectures in the future.
- We will be able to publish a preliminary benchmark of the old vs. new application code, comparing across a selection of hardware that WCG staff have at home. We expect performance improvements and better memory utilization for the new version of the MCM1 and by extension initial implementation of the MAM1 project.
- In light of the merger of this pull request (introducing "BUDA" to the latest BOINC server releases), we find a strong motivation to upgrade our BOINC server version to track BOINC upstream as nearly every bioinformatics application we have experience running in an HPC environment we could run on the grid, if only we could run containers. We have considered multiple different strategies to accomplish this migration in the past, and now that we have occasion to test existing and new applications with the newest BOINC server version during this downtime offsite, we will put together a roadmap after the launch of MAM1 and upgrade BOINC server to use BUDA/containers going forward.
- We are working with the backups of the BOINC and website/forums databases offsite to prepare the Mapping Arthritis Markers project for the launch in January.
- While we have the opportunity, we will look into optimizing the MCM1 and MAM1 applications. One example, we will create an encoding for the fields in the MCM1 configuration file and further compress the already tiny configuration file for MCM1, rather than send it in plain text as before. This update will also be used to streamline the configuration file across individual projects.
- We are also exploring the potential for a GPU version of MCM1 and MAM1 projects, and future applications for searching for biomarker signatures.
- We are working on tiered caching across the download servers where workunits that have been loaded into BOINC recently will trigger a write of the input files to a distributed in-memory cache, backed up by the local disk of the download servers.
- For ARP1 project, we will be merging all file downloads into a single archive. Previously, even the individual files were getting transient HTTP errors/interruptions frequently and had to wait in queue. We did fix that bug before we went down. This should resolve the bug that was interrupting larger downloads and uploads multiple times, and we are also exploring other optimizations that should improve workunit distribution.
- Hopefully, these changes will result in a more efficient file transfers, less I/O pressure on NFS and the storage server, and therefore more throughput under load, which we will need as we look to GPU-enable these and future applications on the grid.
- We are making final preparations to power down. Before powering off all VMs and the storage servers, we will kill all traffic at the load balancer so we can allow the filesystems and databases to settle before taking a final backup.
- When we kill traffic at the load balancer overnight on Dec. 6, there will be no access to the website, forums, BOINC scheduler, BOINC uploads, or BOINC downloads.
- We have transferred database backups for BOINC and the website/forums, as well as all source code offsite. We will be transferring the final backups overnight as well.
- Today (Dec 6), we have been taking snapshots of all production VMs and sometime this evening we should have all instances that make up our backend infrastructure backed up as snapshots, ready to deploy from resilient storage if there are any issues powering up our instances when we are back online in January.
- We will be working on the new project (Mapping Arthritis Markers), long-standing issues with device synchronization between the website and BOINC databases, server status page, real-time stats API, and a new look for the forums as well as new badges during the downtime. We look forward to sharing the results of the data center refresh with you in the New Year - thank you for your support and happy holidays!
- The WCG shutdown date has been moved up from December 9th, 2024 to December 7th, 2024. We were informed by email on November 21st, to shutdown by December 9th because there would be no cooling by that date. Hosting earlier this week asked us to bump the date up to the 7th.
- We have determined it is not feasible to migrate the BOINC infrastructure to another site during downtime, and we are still waiting to hear back from UHN personnel who manage our DNS records to see when we can switch the website and forums over to the alternate site. If we do hear back, this will take place between now and December 6th, 2024.
- As users pointed out in the forums, it did not really make sense to start pushing new ARP1 workunits given the imminent downtime, and we will stop producing new MCM1 workunits tomorrow to hopefully give the bulk of outstanding workunits a chance to be uploaded. When we power down, after all traffic to the upload servers is stopped, deadlines for outstanding workunits will be extended to cover the downtime.
- With the improvements to the cloud environment, we have been informed that the issue with the network agents in our cloud environment causing the website and forums database instance to become inaccessible on the network until hosting intervenes (cause of this past weekend's outage and many previous outages), will be fixed.
- IMPORTANT: We have been notified of an extended downtime at SHARCNET facility for construction lasting from December 9th, 2024 to January 3rd, 2025. There will be no power and no cooling during this time. We are exploring temporary migration to another site. We will provide an update on what downtime if any can be expected to start on December 9th, 2024. Overall, this upgrade should provide further improvements to the WCG capacity.
- Bandwidth has been improved thanks to hosting staff at SHARCNET. In addition, we have more and better hardware devoted to handling downloads and uploads, and a more competent load balancer.
- ARP1 will resume in limited quantities over the next few days. We will make an effort to focus on extremes as suggested in the forums and test the imposed rate limits on workunit production as well as total bandwidth of all clients and number of connections per client for ARP1 file transfer specifically.
- Forums were down earlier today for an extended period, they are back up now and we apologize for the slow response.
- SHARCNET is tuning performance on the main network node for our cloud environment, users may have noticed some service interruption today as a result.
- We have identified a way to increase bandwidth to the expected level in our cloud environment, which if today's testing correlates will offer at least an order of magnitude improvement and hopefully more.
- However, the approach identified will require updates to DNS records hosted by a separate team at UHN and more investigation to confirm feasibility. We will update volunteers when we have firm timelines and if there will be any downtime when we switch DNS over; assuming we do not find a simpler solution that requires no change to DNS records before verifying this is the way to go with SHARCNET's help.
- With the additional hosts we have been able to provision and the local disk available to them, we are hopeful that we can more intelligently write input files and output files to local disk based on a modulo of the workunit ID rather than relying on NFS for everything - the local disks are RAID1 SSDs with ~1TB available for caching of downloads in this way and potentially uploads to researchers that never need to touch NFS for more than metadata. This effort will take some time, but we expect to make reasonable progress towards this goal.
- Working with hosting staff today, we confirmed that the network in our cloud environment is not behaving as expected, the congestion and latency do not make sense at this bandwidth or looking at the number of packets in flight at any given time.
- We should expect a large increase in available bandwidth when these issues are resolved, where we though it was only server resources limiting transfer quantity and rates, therefore causing 503s which translated as "Transient HTTP error" in the BOINC client.
- Hosting at SHARCNET were able to recommend some kernel settings they often use for hosts fielding many connections, and also change some offload settings on their end to help alleviate the problem while we pursue a full resolution.
- With the above changes, we have seen a higher maximum throughput at the load balancer, typically averaging over 100Mbps wheras before the average was under with ~100Mbps spikes, spikes are now hitting ~150Mbps.
- ARP1 was soft-paused last week, there are only 361 left to claim as of this writing.
- Approximately ~29,000 ARP1 workunits must now be uploaded, before we can titrate ARP1 slowly to the right level, after implementing traffic shaping and rate limits in workunit creation, download, and upload.
- Deadlines for ARP1 workunits have been extended again on Saturday Nov 16th, 2024, by 5 days. We will continue to extend the deadline of ARP1 workunits.
- We fixed bugs in the HAProxy and Apache configuration files that were causing connection resets on large file transfers, we hope users have seen that despite the atrocious speeds large file transfers are more likely to succeed. ARP1 should download a single file and upload a single file. We will work on this.
- Traffic shaping and ratelimits will be applied at the load balancer and backend webservers to so that connection slots are less available to client IP addresses specifically for ARP1 file transfers, and the overall bandwidth of all ARP1 transfers is always limited to a reasonable fraction of total available bandwidth.
- We provisioned 3 new servers, more to come, with the help of SHARCNET. They have much more CPU, memory, and even a large local disk.
- We are pursuing tiered caching of files for download during creation of the workunit to bulk transfer or write directly to local disk of download servers. We will also explore in-memory caching especially for smaller files, we believe this will reduce load on NFS, SAN, storage server, increase available bandwidth server availability and therefore throughput
- We migrated all provisioning scripts, code and configuration, as well as build and deploy scripts previously supporting only CentOS 7 to support also Ubuntu 22 as that was the only guest OS we could run on this new server group provisioned by SHARCNET.
- We migrated the production load balancer, upgrading HAProxy from v1.8 -> v2.8, to one of these servers. We deployed two new download servers into production in this way, and pending a final fussing with builds that work on CentOS 7 but not yet Ubuntu 22 we will be able to deploy the binaries required to have them accept file uploads as well. We tuned kernel parameters and application configuration files especially HAProxy and HTTPD/Apache2 with some napkin math for these new servers and the existing older CentOS 7 servers as well. With the massive increase in CPU, memory, and understanding of what does what in each configuration file resulting in optimizations, we are still only doing ~100Mbps in total in our best moments at any given time through the production load balancer. Therefore, it seems we are ultimately bandwidth limited at the moment, likely a combination of low available bandwidth and inefficient use of that bandwidth.
- Increasing the maxconn and timeout queue of the new download servers to throw less 503 errors, still resulted in timeouts, but also made all aspects of the network in our environment unuseable due to latency. Users may have noticed in their BOINC clients that at some points over the last two weeks high timeout queue settings and aggressive keep-alive settings would cause retries in the BOINC client to appear to be stuck, not backing off for minutes in some cases, only to be eventually rejected and only rare cases start late finally pushing through the queue to connect through to the backend. We were experimenting to see if we could serve more requests by having them queue up for longer before being hit with a 503 Service Unavailable, but in those cases, as in many other regimes we found that throughput barely increases while the network congestion/latency renders the entire cloud environment unuseable. The website, my terminal, CI/CD jobs, git repos, same or different subnet. We have been aggressively testing in prod and that is bad practice, but we only pushed and evaluated configurations that we believed held promise to improve the situation in some way. We apologize for the chaos and confusion.





