
Home
About Us
Description
Updates
Mapping Cancer Markers
Towards Precision Medicine
Towards Precision Medicine
- Highlights
- the Top 100 AI in Oncology leaders list,
(Deep Knowledge Analytics) - Ganatra D, Kotlyar M, Dohey A, Codner D, Li Q, Abji F, Rasti M, Eder L, Gladman D, Rahman P, Jurisica I, Chandran V. Combining Clinical, Genetic and Protein Markers Using Machine Learning Models Discriminates Psoriatic Arthritis Patients From Those With Psoriasis. J Psoriasis Psoriatic Arthritis. 24755303251344134. doi: 10.1177/24755303251344134, 2025. Epub ahead of print.
- Yuan Z, Ostrowska-Podhorodecka Z, Cox T, Norouzi M, Wang Y, Robaszkiewicz K, Siatkowska M, Xia K, Ali A, Abovsky M, Jurisica I, Smith P, McCulloch CA. Annexin A2 Contributes to Release of Extracellular Vimentin in Response to Inflammation. FASEB J, 39(9):e70621, 2025.
- Dang S, Li X, Diao L, Piguet V, Croitoru D, Wither J, Jurisica I, Chandran V, Eder L. Sex differences in serum proteomic profiles in psoriatic arthritis. Rheumatology (Oxford). keaf311, 2025. Epub ahead of print.
- Liao Z, Kumar K, Kopal J, Huguet G, Saci Z, Jean-Louis M, Pausova Z, Jurisica I, Bearden CE; IMAGEN Consortium; 16p11.2 European Consortium; Jacquemont S, Paus T. Copy number variants and the tangential expansion of the cerebral cortex. Nat Commun. 16(1):1697, 2025.
Cancer development is a multi-step process that leads to uncontrolled tumour cell growth caused by and resulting in complex changes: many genes are amplified, deleted, mutated, up- or down-regulated; many proteins and pathways are activated or suppressed. Estimating across 1.9 million patients from 31 countries and 5 continents, current treatments achieve a 5-year survival rate for less than 50% of diagnosed cancer (Coleman et al. Cancer survival in five continents: a worldwide population-based study (CONCORD). Lancet Oncol 9(8): 730-756, 2008).
Years of research improved survival in breast and prostate cancers by finding molecular markers for early diagnosis and by individualized treatment. However, pancreatic cancer remains almost 100% lethal, and the overall survival rate for lung cancer has improved barely during the past decades, having only moved from 13% to 16%.
The Mapping Cancer Markers (MCM) project aims to comprehensively and systematically discover clinically useful markers to aid early cancer detection, identification of high-risk patients, and prediction of treatment response.
To power this research, we rely on World Community Grid volunteers who donate their computers' spare capacity to carry out this extensive analysis. Finding all clinically useful markers would require processing thousands of patient samples and testing an astronomical number of marker combinations, which is not feasible even on World Community Grid. Instead, we use heuristics to reduce the search space, enabling us to tackle this challenge with the computing resources donated by volunteers like you.
Support our research and join World Community Grid today!
Thank you for your support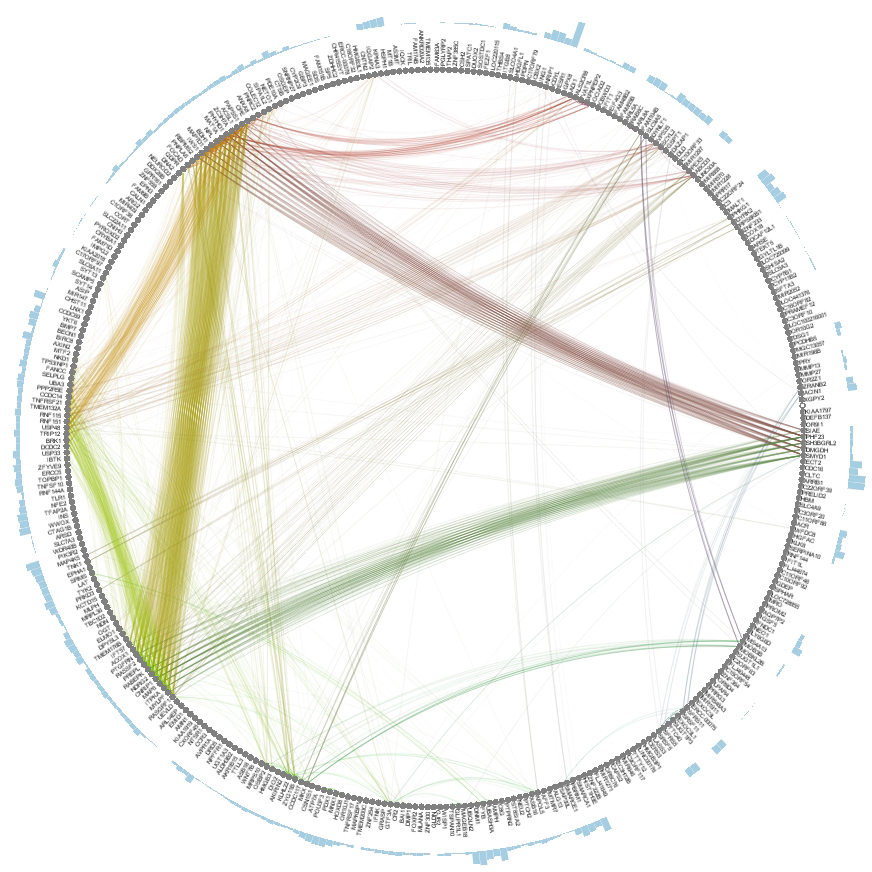
Years of research improved survival in breast and prostate cancers by finding molecular markers for early diagnosis and by individualized treatment. However, pancreatic cancer remains almost 100% lethal, and the overall survival rate for lung cancer has improved barely during the past decades, having only moved from 13% to 16%.
The Mapping Cancer Markers (MCM) project aims to comprehensively and systematically discover clinically useful markers to aid early cancer detection, identification of high-risk patients, and prediction of treatment response.
To power this research, we rely on World Community Grid volunteers who donate their computers' spare capacity to carry out this extensive analysis. Finding all clinically useful markers would require processing thousands of patient samples and testing an astronomical number of marker combinations, which is not feasible even on World Community Grid. Instead, we use heuristics to reduce the search space, enabling us to tackle this challenge with the computing resources donated by volunteers like you.
Support our research and join World Community Grid today!
Thank you for your support


The original MCM team
Current team members

Igor Jurisica
Principal Investigator
Scientific Director
Principal Investigator
Scientific Director

Dylan Bethune-Waddell
Data Science Specialist
Data Science Specialist

Chiara Pastrello
Research Associate / Lab Manager
Research Associate / Lab Manager

Max Kotlyar
Research Associate
Research Associate

Dr. Serene Wong
Research Associate
Research Associate

Richard Lu
SA/DBA/Research Programmer
SA/DBA/Research Programmer

Dr. Mark Abovsky
Applications and Web Specialist
Applications and Web Specialist

Michelle Morgovsky
WCG Communications intern
WCG Communications intern

Gabriel Gonzales Vargas
WCG Communications intern
WCG Communications intern
Cancer Challenge
Cancer development is a multi-step process that leads to uncontrolled tumour cell growth caused by and resulting in complex changes: many genes are amplified, deleted, mutated, up- or down-regulated, many pathways are activated or suppressed.Estimating from the CONCORD study across 1.9 million patients from 31 countries and 5 continents, current treatments achieve a 5-year survival rate for less than 50% of diagnosed cancer (Coleman, 2008). Approximately 12.7 million new cancer cases were diagnosed worldwide in 2008. Lung, female breast, colorectal & stomach cancers account for more than 40% of all cases. Almost 8 million deaths from cancer occurred worldwide in 2008. Lung, stomach, liver, colorectal & female breast cancers account for more than 50% of all cancer deaths (http://www.cancerresearchuk.org/cancer-info/cancerstats/world/).
Canadian cancer statistics (2012) indicates that lung cancer accounts for almost 14% of all cancer cases in Canada, leading to the highest number of deaths (20,100, 27% of all cancers). Years of research improved relative survival by 6% for all cancers since 1992. Highest improvements are for non-Hodgkin lymphoma & leukemias. Success in breast and prostate cancers has been achieved by finding molecular markers for early diagnosis and by individualized treatment. Highest survival is for thyroid, prostate & testicular cancers. However, pancreatic cancer remains almost 100% lethal, and the overall survival rate for lung cancer has improved barely during the past decades, having only moved from 13% to 16%.
Cancer is caused by genetic changes or environmental effects that interfere with the mechanisms that control cell growth. These changes, as well as normal cell activities, can be detected in tissue samples through the presence of unique indicators, such as DNA and proteins, which together are known as “markers.” Specific combinations of these markers may be associated with a given type of cancer, indicate patient's risk of cancer recurrence, or indicate probability of patient's response to treatment. While several markers are already known to be associated with certain cancers, there are many more to be discovered, as cancer is highly heterogeneous.
The discovery and validation of biomarkers is complex and computationally intensive process. It involves analyzing hundreds of thousands of parameters (clinical variables, gene, protein, microRNA, activity, etc.) to identify subsets that best describe patients, their prognosis and response to treatment. Finding all clinically useful markers and selecting the best subset represents a challenging computational optimization task as we would need to compare all possible parameter combinations.
Canadian cancer statistics (2012) indicates that lung cancer accounts for almost 14% of all cancer cases in Canada, leading to the highest number of deaths (20,100, 27% of all cancers). Years of research improved relative survival by 6% for all cancers since 1992. Highest improvements are for non-Hodgkin lymphoma & leukemias. Success in breast and prostate cancers has been achieved by finding molecular markers for early diagnosis and by individualized treatment. Highest survival is for thyroid, prostate & testicular cancers. However, pancreatic cancer remains almost 100% lethal, and the overall survival rate for lung cancer has improved barely during the past decades, having only moved from 13% to 16%.
Cancer is caused by genetic changes or environmental effects that interfere with the mechanisms that control cell growth. These changes, as well as normal cell activities, can be detected in tissue samples through the presence of unique indicators, such as DNA and proteins, which together are known as “markers.” Specific combinations of these markers may be associated with a given type of cancer, indicate patient's risk of cancer recurrence, or indicate probability of patient's response to treatment. While several markers are already known to be associated with certain cancers, there are many more to be discovered, as cancer is highly heterogeneous.
The discovery and validation of biomarkers is complex and computationally intensive process. It involves analyzing hundreds of thousands of parameters (clinical variables, gene, protein, microRNA, activity, etc.) to identify subsets that best describe patients, their prognosis and response to treatment. Finding all clinically useful markers and selecting the best subset represents a challenging computational optimization task as we would need to compare all possible parameter combinations.
Approach
The Mapping Cancer Markers (MCM) project focuses on clinical application - discovering specific groups of markers that can be used to improve detection, diagnosis, prognosis and treatment of cancer. As a second goal, the comprehensive analysis of existing molecular profiles of cancer samples will lead to unraveling characteristics of such groups of markers - and in turn improving our understanding how to find them more efficiently.
Our strategy to reduce mortality includes three steps:
Our strategy to reduce mortality includes three steps:
- Increase number of cases diagnosed at earlier stage
- We need to identify biomarkers for early cancer detection
- Individualized treatment
- We need to find biomarkers for treatment selection and response monitoring
- Improved treatment
- We need to improve our understanding of disease mechanism and drug mechanism of action
- We need to identify useful drug combinations and design new medicines
References
- Coleman, M. P., M. Quaresma, et al. (2008). "Cancer survival in five continents: a worldwide population-based study (CONCORD)." Lancet Oncol 9(8): 730-756.
- Ein-Dor, L., I. Kela, et al. (2005). "Outcome signature genes in breast cancer: is there a unique set?" Bioinformatics 21(2): 171-178.
- Ein-Dor, L., O. Zuk, et al. (2006). "Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer." Proc Natl Acad Sci U S A 103(15): 5923-5928.

July 2025 Update
We continue to investigate molecular signatures linked to lung cancer, with a current focus on ECD (Ecdysoneless Cell Cycle Regulator), a gene that regulates cell growth and repair.
Read More
We continue to investigate molecular signatures linked to lung cancer, with a current focus on ECD (Ecdysoneless Cell Cycle Regulator), a gene that regulates cell growth and repair.
Read More

May 2025 Update
We continue to investigate molecular signatures linked to lung cancer, with a current focus on PDE8B (Phosphodiesterase 8B), a gene that is involved in hormone synthesis and has altered expression in diverse cancers.
Read More
We continue to investigate molecular signatures linked to lung cancer, with a current focus on PDE8B (Phosphodiesterase 8B), a gene that is involved in hormone synthesis and has altered expression in diverse cancers.
Read More

March 2025 Update
We continue to investigate molecular signatures linked to lung cancer, with a current focus on DYNLT1 (Dynein Light Chain Tctex-Type 1), a gene with pivotal roles in intracellular transport and cancer pathophysiology.
Read More
We continue to investigate molecular signatures linked to lung cancer, with a current focus on DYNLT1 (Dynein Light Chain Tctex-Type 1), a gene with pivotal roles in intracellular transport and cancer pathophysiology.
Read More

January 2025 Update
We continue to investigate molecular signatures linked to lung cancer, with a current focus on GCM1, a transcription factor involved in placental development and potentially significant in lung cancer biology. The GCM1 gene encodes a protein with a gcm-motif, playing a central role in DNA-binding and regulating placental-specific genes. While its primary function is in placental biology, emerging evidence suggests GCM1 mutations are implicated in various cancers, including lung adenocarcinoma (ADC).
Read More
We continue to investigate molecular signatures linked to lung cancer, with a current focus on GCM1, a transcription factor involved in placental development and potentially significant in lung cancer biology. The GCM1 gene encodes a protein with a gcm-motif, playing a central role in DNA-binding and regulating placental-specific genes. While its primary function is in placental biology, emerging evidence suggests GCM1 mutations are implicated in various cancers, including lung adenocarcinoma (ADC).
Read More

July 2024 Update
We continue to characterise lung cancer biomarkers identified in the MCM1 project. This update focuses on ASTN2, a protein involved in neuronal migration. It is expressed across several tissue types, and has been implicated in various cancers.
Read More
We continue to characterise lung cancer biomarkers identified in the MCM1 project. This update focuses on ASTN2, a protein involved in neuronal migration. It is expressed across several tissue types, and has been implicated in various cancers.
Read More

May 2024 Update
We continue to characterise lung cancer biomarkers identified in the MCM1 project. This update focuses on KLF5, a well-studied Krueppel-like factor 5 transcription factor that regulates several of our identified genes and has a protective role in lung cancer.
Read More
We continue to characterise lung cancer biomarkers identified in the MCM1 project. This update focuses on KLF5, a well-studied Krueppel-like factor 5 transcription factor that regulates several of our identified genes and has a protective role in lung cancer.
Read More

March 2024 Update
We continue to characterise lung cancer biomarkers identified in the MCM1 project. This update focuses on HSD17B11, a gene associated with lung cancer survival. HSD17B11 is a protein coding gene, relatively ubiquitously expressed across organs and tissues. It is a short-chain alcohol dehydrogenase that metabolises secondary alcohols and ketones.
Read More
We continue to characterise lung cancer biomarkers identified in the MCM1 project. This update focuses on HSD17B11, a gene associated with lung cancer survival. HSD17B11 is a protein coding gene, relatively ubiquitously expressed across organs and tissues. It is a short-chain alcohol dehydrogenase that metabolises secondary alcohols and ketones.
Read More
February 2024 Update
We continue our work on characterizing lung cancer biomarkers identified in the MCM1 project. This update focuses on a transcriptional corepressor TLE3.
Read More
We continue our work on characterizing lung cancer biomarkers identified in the MCM1 project. This update focuses on a transcriptional corepressor TLE3.
Read More
January 2024 Update
We continue our work on characterizing lung cancer biomarkers identified in the MCM1 project. This update focuses on PCSK5, a gene associated with lung cancer survival and which shows differential expression across various cancer types.
Read More
We continue our work on characterizing lung cancer biomarkers identified in the MCM1 project. This update focuses on PCSK5, a gene associated with lung cancer survival and which shows differential expression across various cancer types.
Read More
November 2023 Update
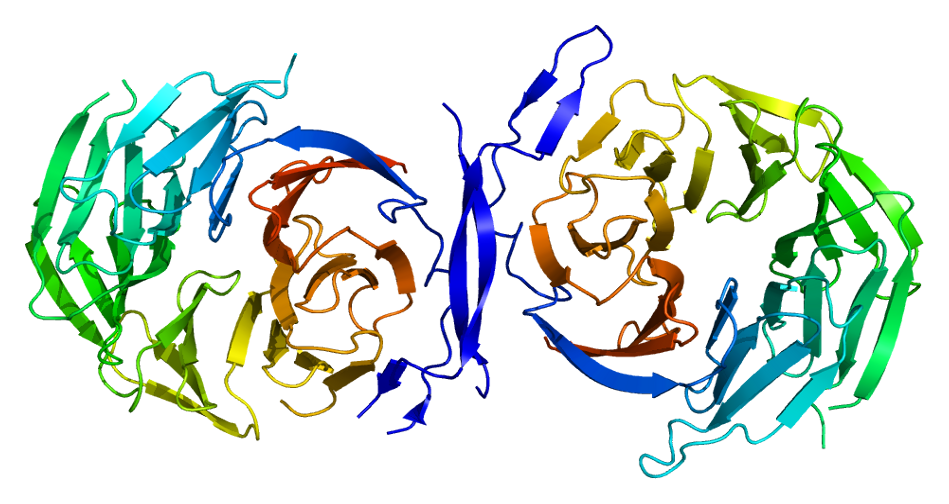
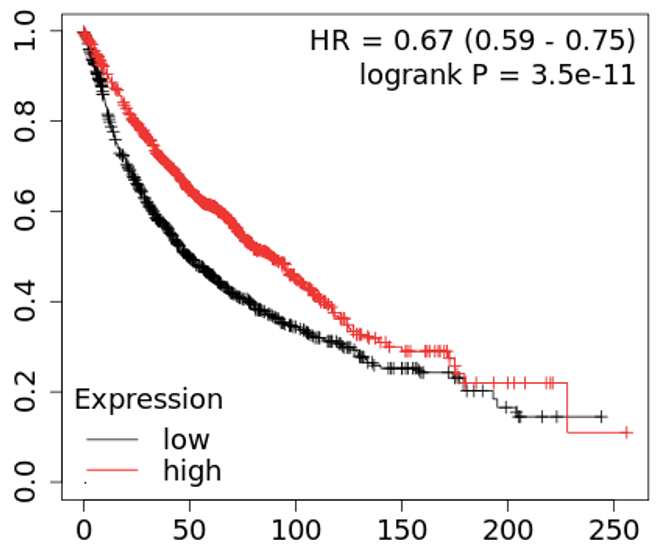
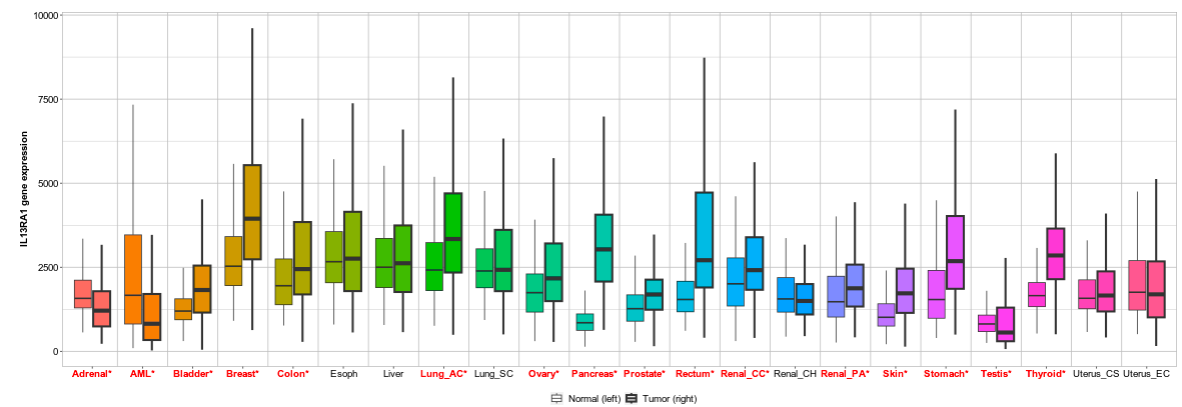
We continue our work on characterizing lung cancer biomarkers identified in the MCM1 project. This update focuses on IL13RA1, a gene associated with lung cancer survival and differentially expressed across multiple cancer types compared to normal tissues.
Read More
We continue our work on characterizing lung cancer biomarkers identified in the MCM1 project. This update focuses on IL13RA1, a gene associated with lung cancer survival and differentially expressed across multiple cancer types compared to normal tissues.
Read More
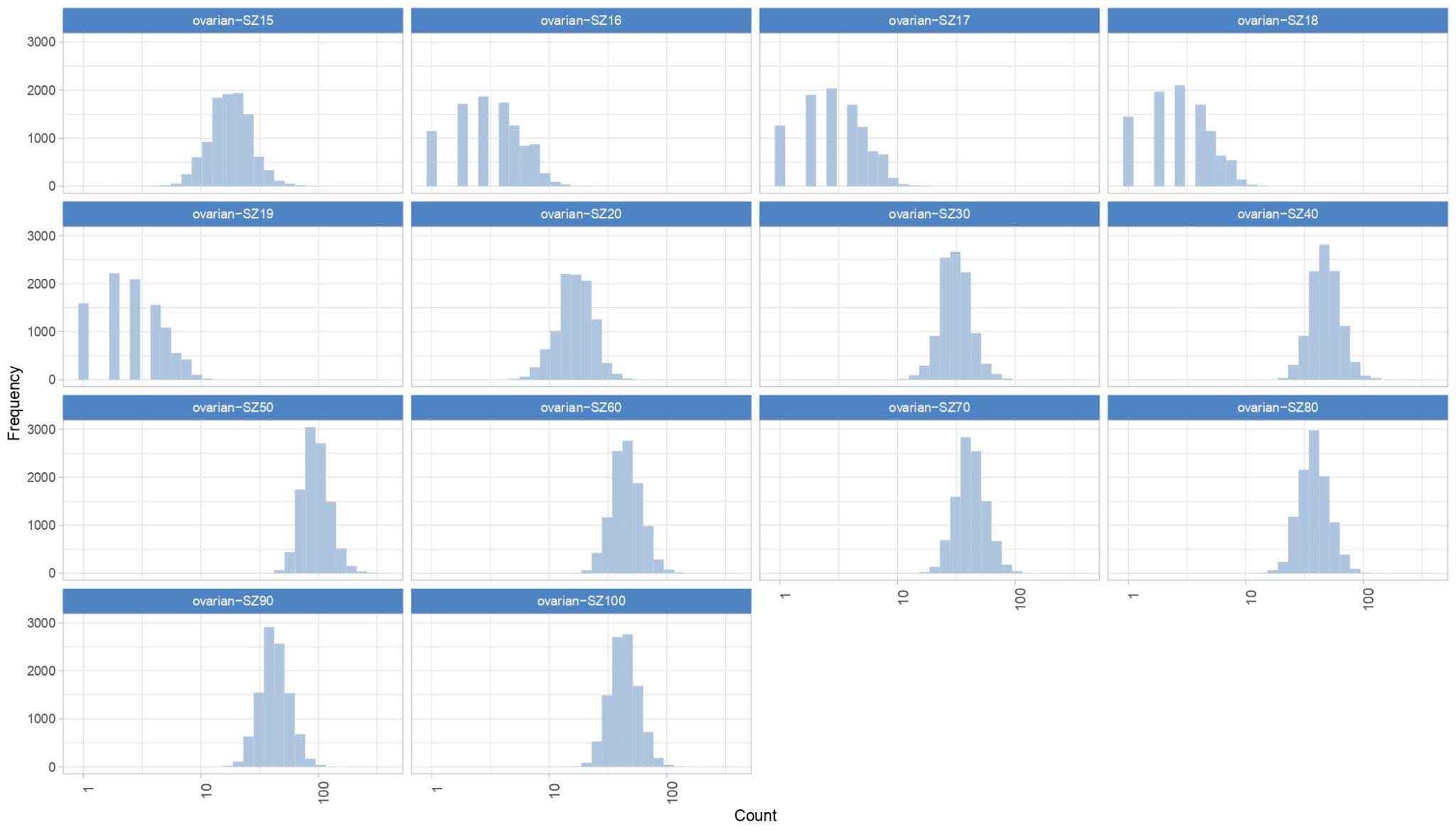
September 2023 Update
Since the ovarian cancer results were dishomogeneous, we will begin running additional ovarian work units starting September 25th.
Read More
Since the ovarian cancer results were dishomogeneous, we will begin running additional ovarian work units starting September 25th.
Read More
September 2023 Update
We continue our work on characterizing lung cancer biomarkers identified in the MCM1 project. This update focuses on ADH6, a gene associated with smoking status and lung cancer prognosis.
Read More
We continue our work on characterizing lung cancer biomarkers identified in the MCM1 project. This update focuses on ADH6, a gene associated with smoking status and lung cancer prognosis.
Read More
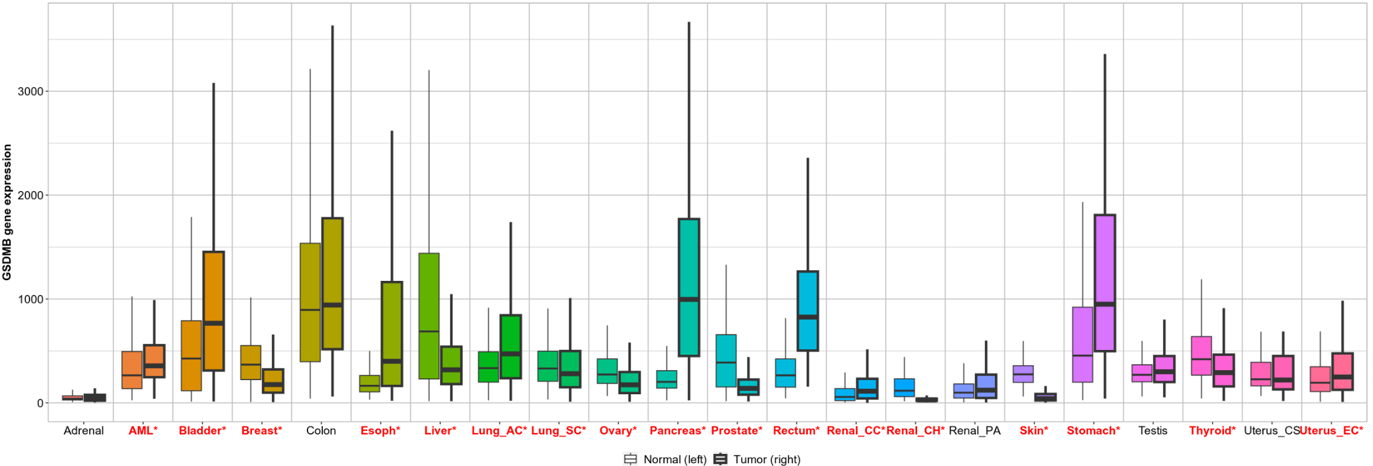
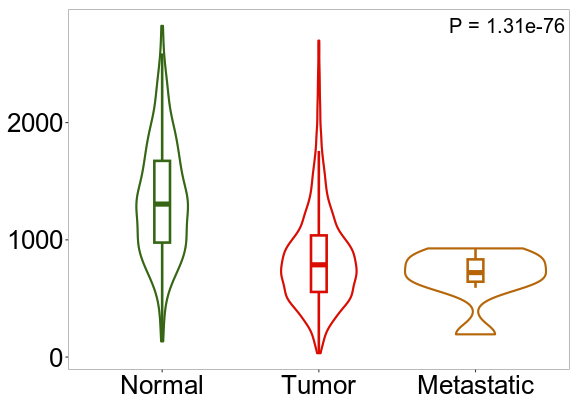
July 2023 Update
We continue our work on characterizing lung cancer biomarkers identified in the MCM1 project. This update focuses on GSDMB, a gene associated with lung cancer survival and differentially expressed across multiple cancer types compared to normal tissues.
Read More
We continue our work on characterizing lung cancer biomarkers identified in the MCM1 project. This update focuses on GSDMB, a gene associated with lung cancer survival and differentially expressed across multiple cancer types compared to normal tissues.
Read More
April 2023 Update
Continuing research into putative lung cancer biomarkers, we have identified 26 genes that are present with top scores across all the signature sizes considered. This update focuses on a gene called FARP1, which is linked to lung cancer metastasis.
Read More
Continuing research into putative lung cancer biomarkers, we have identified 26 genes that are present with top scores across all the signature sizes considered. This update focuses on a gene called FARP1, which is linked to lung cancer metastasis.
Read More
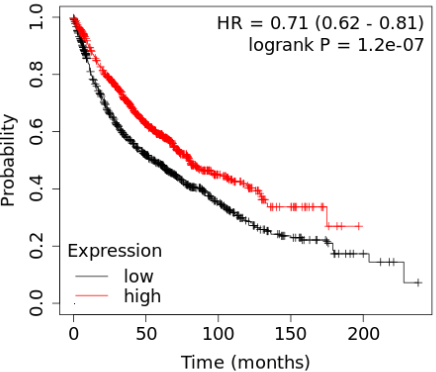
March 2023 Update
We have identified 26 genes that are present with top scores across all the signature sizes considered. This update focuses on VAMP1, a gene linked to patient survival and differentially expressed in normal lung compared to lung cancer.
Read More
We have identified 26 genes that are present with top scores across all the signature sizes considered. This update focuses on VAMP1, a gene linked to patient survival and differentially expressed in normal lung compared to lung cancer.
Read More
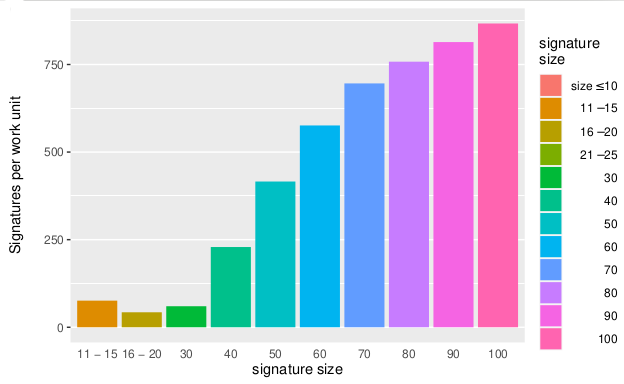
November 2021 Update
Volunteers tested 15 trillion signatures for Mapping Cancer Markers project.
Read More
Volunteers tested 15 trillion signatures for Mapping Cancer Markers project.
Read More
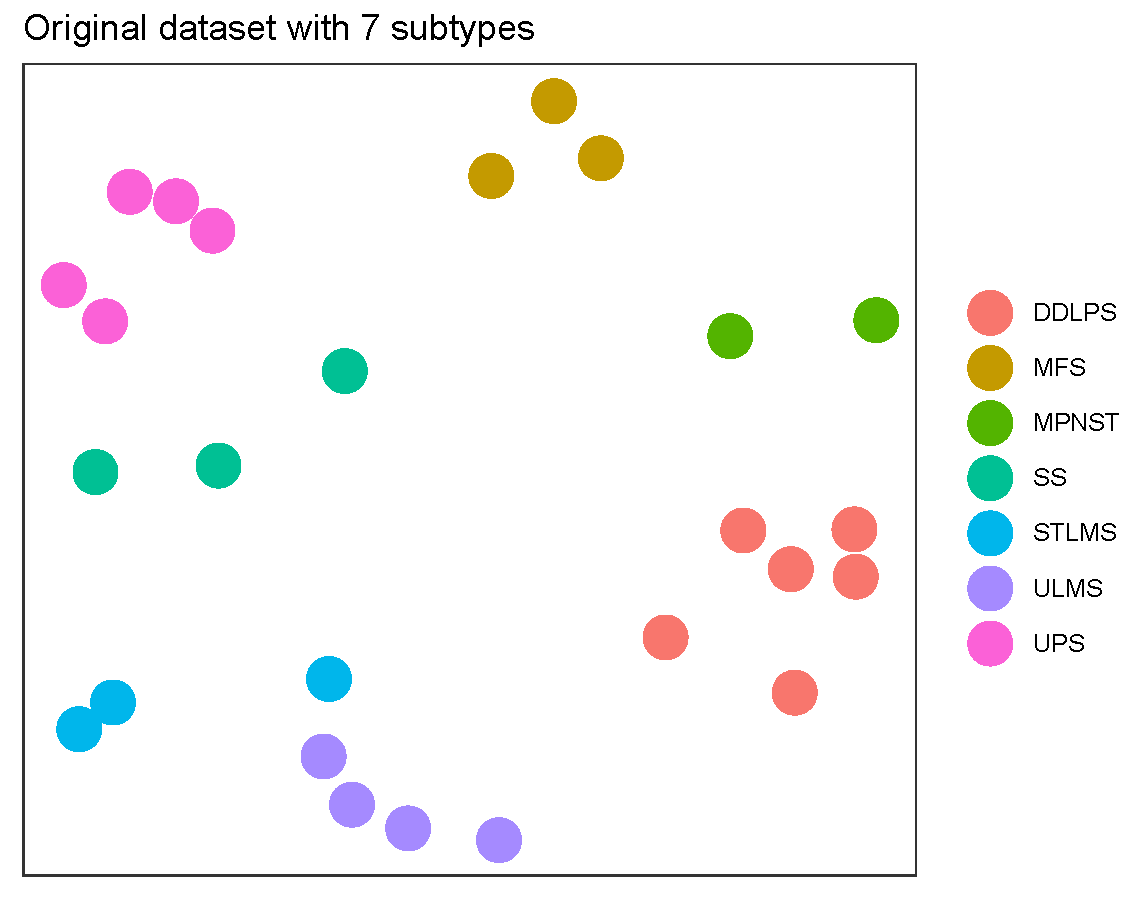
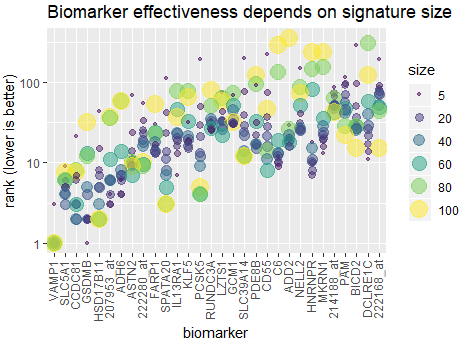
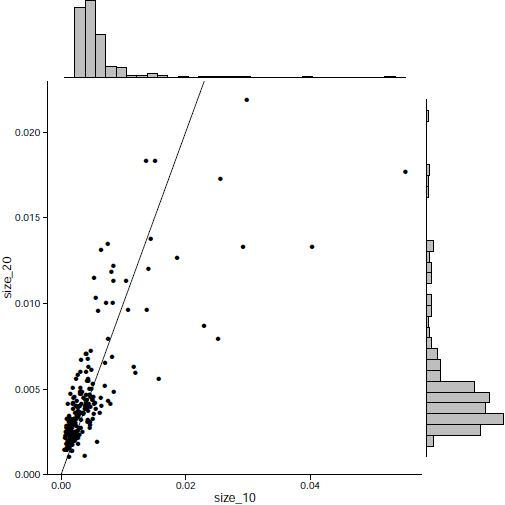
December 2019 update
The effectiveness of each biomarker depends on the signature size, affecting each biomarker differently.
Read More
The effectiveness of each biomarker depends on the signature size, affecting each biomarker differently.
Read More
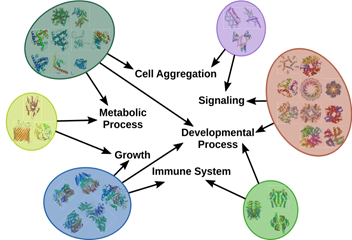
July 2018 update
Multiple groups of biomarkers exist primarily due to redundancy and complex wiring of the biological system.
Read More
Multiple groups of biomarkers exist primarily due to redundancy and complex wiring of the biological system.
Read More

November 2017 update
After 17+ years at Ontario Cancer Institute (now Princess Margaret Cancer Centre) we have joined Krembil Research Institute (KRI) to work on a more complex approach to chronic diseases.
Read More
After 17+ years at Ontario Cancer Institute (now Princess Margaret Cancer Centre) we have joined Krembil Research Institute (KRI) to work on a more complex approach to chronic diseases.
Read More
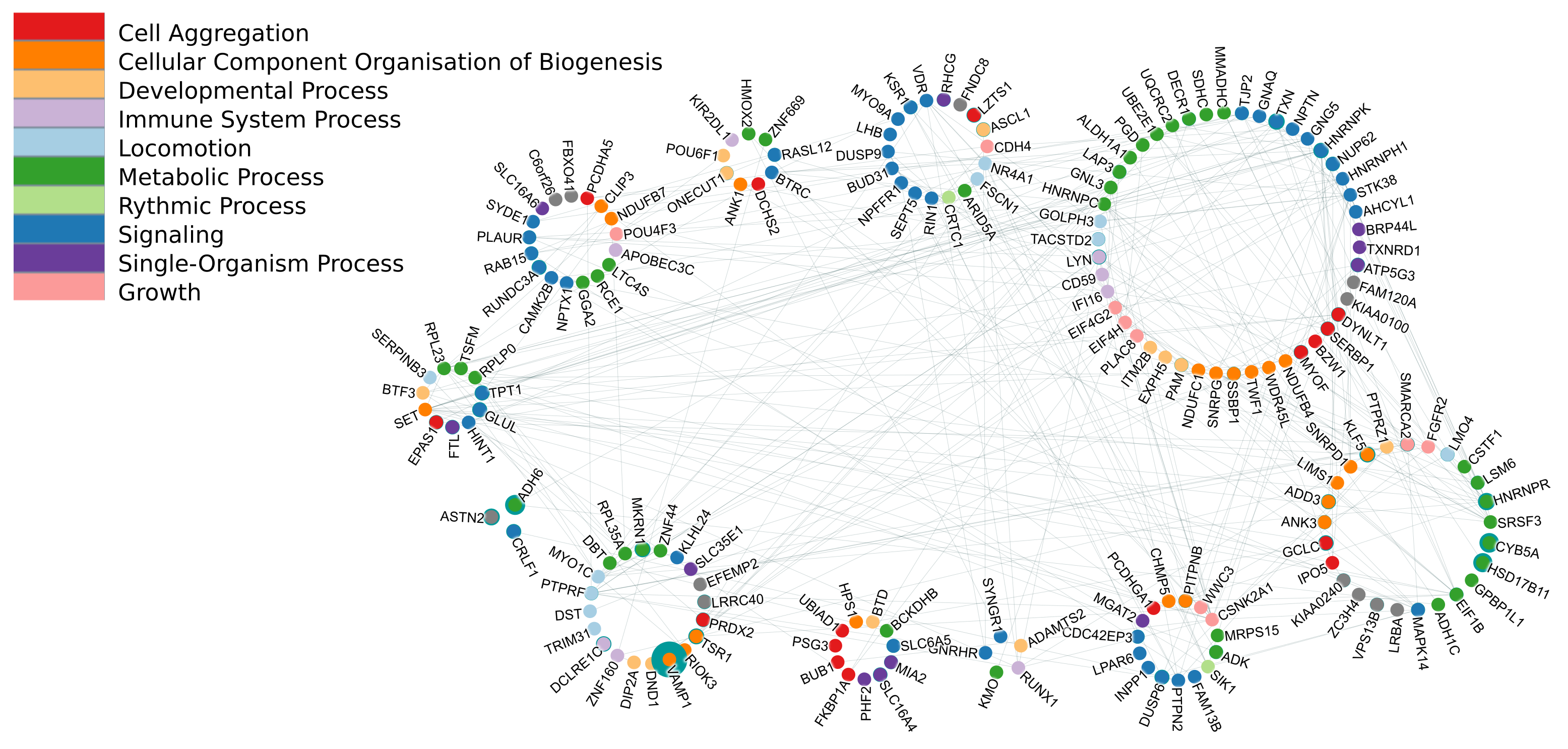
March 2017 update
Characterizing 45 million high-performing signatures derived from World-Community-Grid-computed MCM results.
Read More
Characterizing 45 million high-performing signatures derived from World-Community-Grid-computed MCM results.
Read More
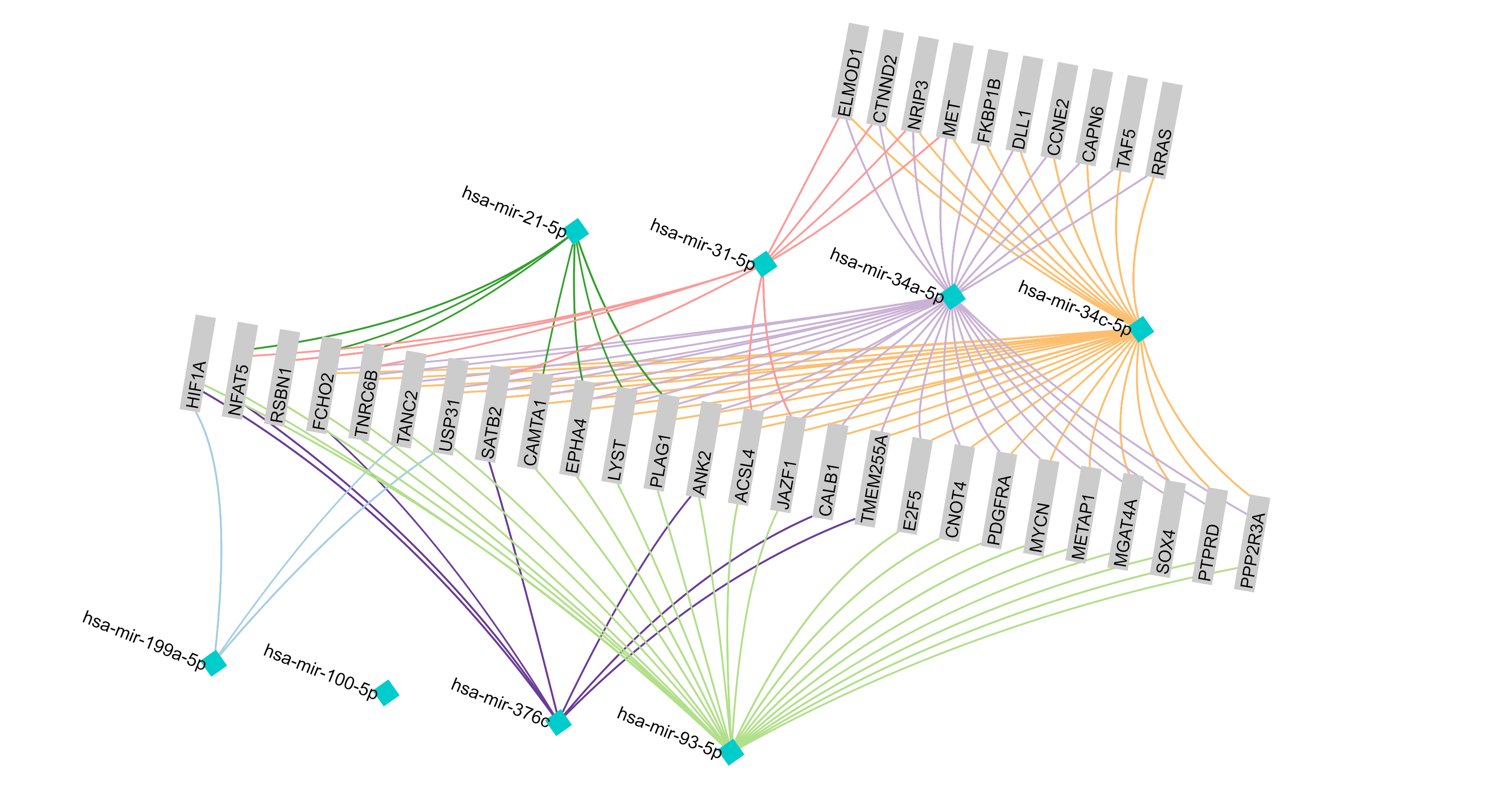
September 2016 update
Exploring ovarian cancer-associated miRNAs and genes to decipher possible regulatory mechanism.
Read More
Exploring ovarian cancer-associated miRNAs and genes to decipher possible regulatory mechanism.
Read More
November 2015 update
Third phase of lung cancer analysis underway: targeting high-scoring, uncorrelated biomarkers.
Read More
Third phase of lung cancer analysis underway: targeting high-scoring, uncorrelated biomarkers.
Read More
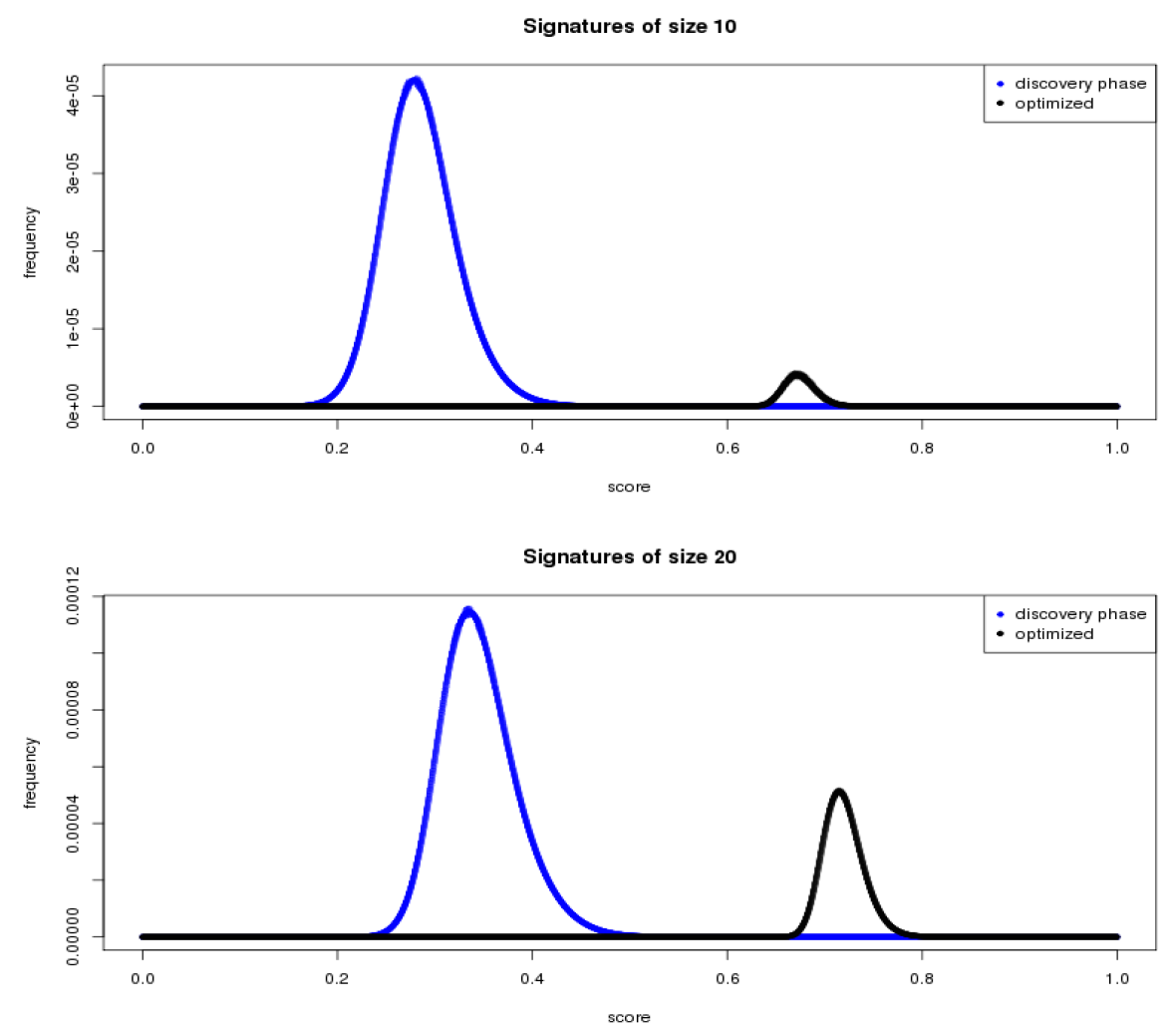
June 2015 update
We used results from the first phase to narrow the field of potential biomarkers from 22,000+ to a subset of 223.
Read More
We used results from the first phase to narrow the field of potential biomarkers from 22,000+ to a subset of 223.
Read More
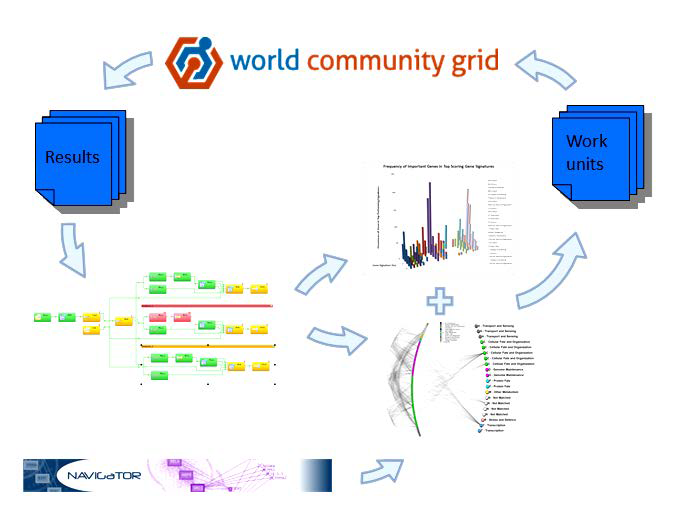
February 2015 update
Starting a gradual and seamless transition to the new phase of MCM, with no interruption in the supply of work units, and no changes to the visualization or code.
Read More
Starting a gradual and seamless transition to the new phase of MCM, with no interruption in the supply of work units, and no changes to the visualization or code.
Read More
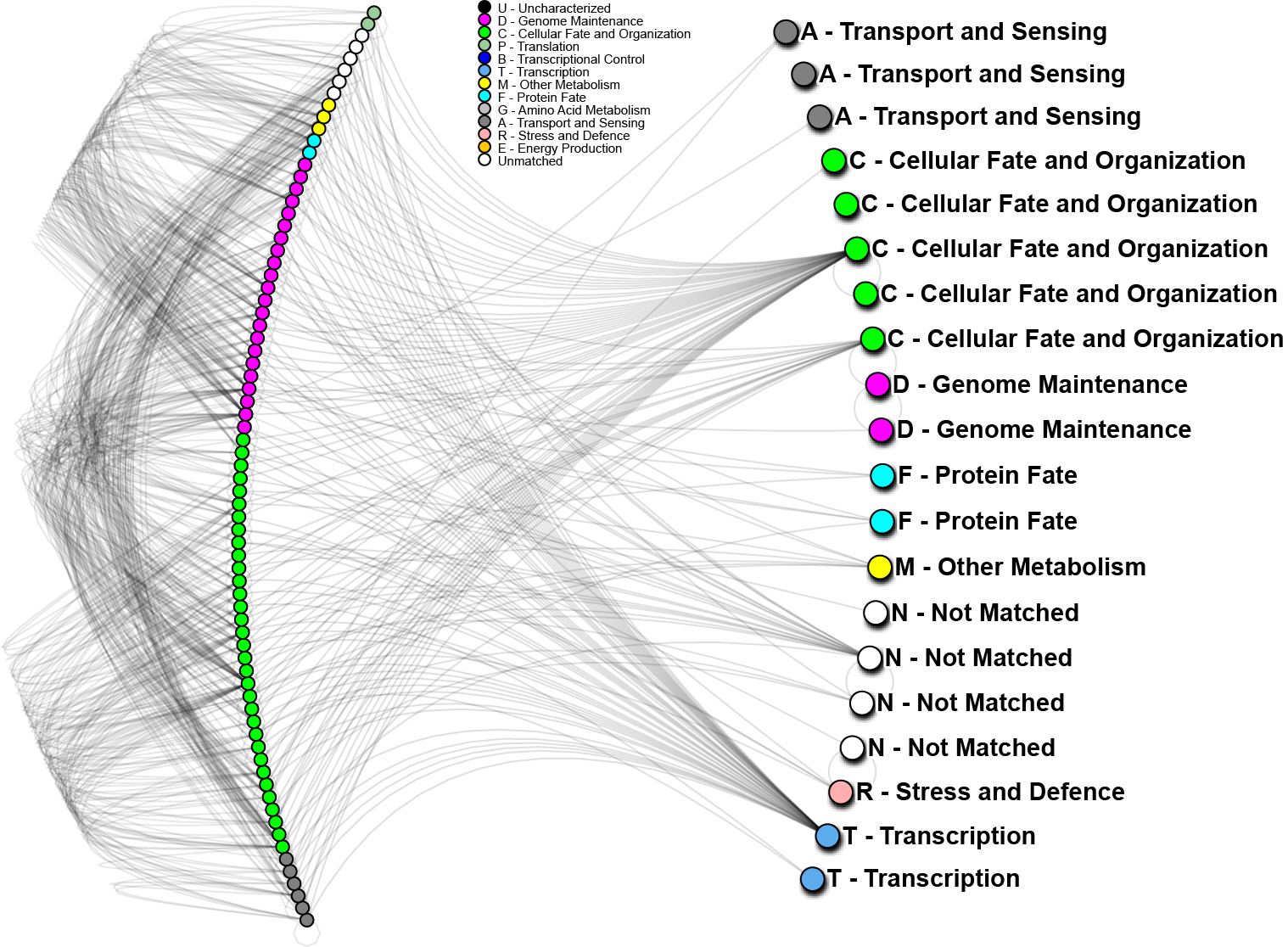
October 2014 update
To further analyze the nature of our top performing genes, we can identify their inter-relations in biological networks.
Read More
To further analyze the nature of our top performing genes, we can identify their inter-relations in biological networks.
Read More






