
Ruian (Ian) Shi
PhD Candidate in Computer Science at the University of Toronto
I am a PhD Candidate at the University of Toronto’s Department of Computer Science supervised by Quaid Morris and Rahul Krishnan.
I'm currently working to improve self-supervised approaches to genomic foundation models. My other research interests involve generative modelling, deep time series modelling, causal inference, and applications in health and biology.
Previously, I completed my BSc and MSc at UofT in CS and worked as an SDE at Amazon. In recent summers, I was at Amazon as an Applied Scientist Intern working on grocery demand forecasting and at Pinterest Labs as a Reseach Science Intern working on NER.
Publications
* indicates equal contribution. † indicates equal senior authorship.
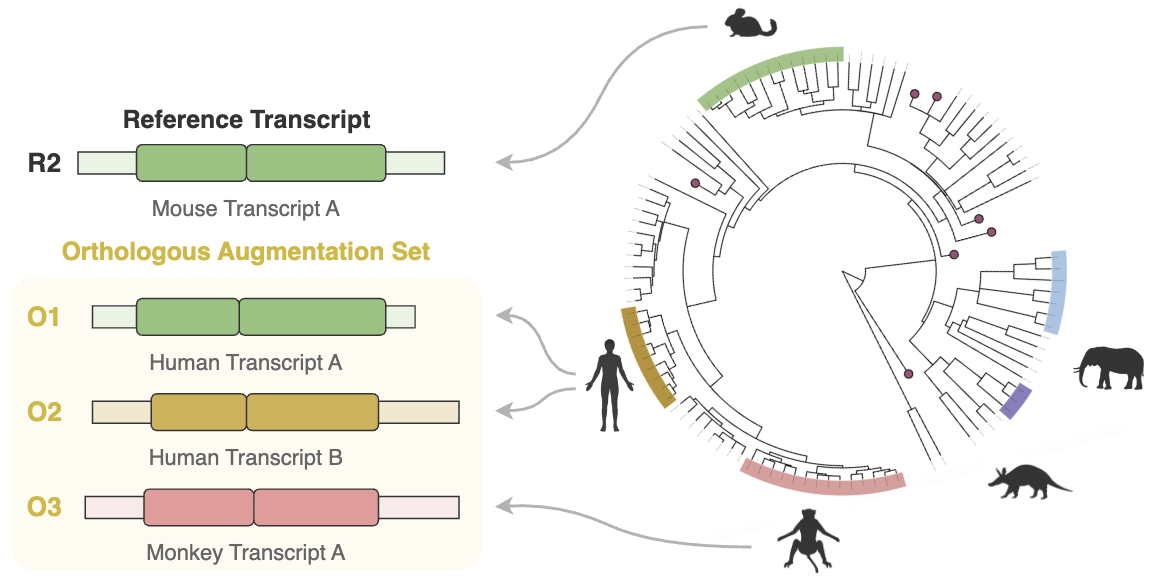
Orthrus: Towards Evolutionary and Functional RNA Foundation ModelsWe developed Orthrus, an RNA foundation model pre-trained with a novel contrastive learning approach incorporating biological augmentations. Using curated RNA transcript pairs from splice isoforms and orthologous genes, Orthrus generates latent representations that cluster sequences by functional and evolutionary similarity. Orthrus represents more than 45 million RNA transcripts and 850 million pairwise interactions. Orthrus outperforms existing genomic models on five mRNA property prediction tasks, requiring minimal fine-tuning data, and can uniquely disambiguiate isoform-specific function. Preprint, In submission
Presented as talks at ISMB2024, MLCB2024. Poster at GEM Workshop @ ICLR 2023 Spotlight at AIDrugX Workshop @ Neurips 2024. |
|
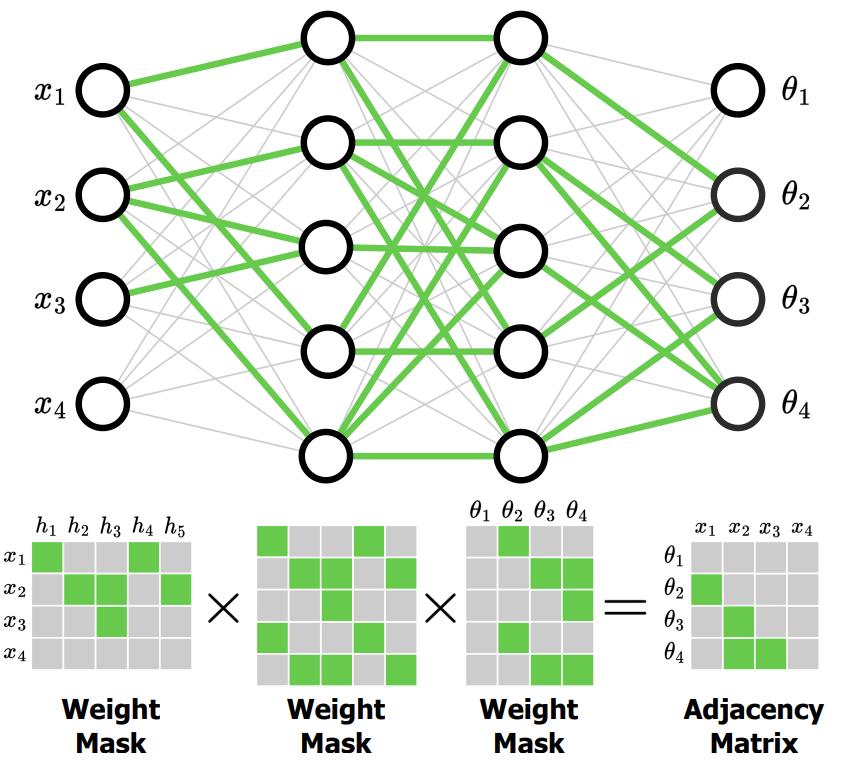
Structured Neural Networks for Density Estimation and Causal InferenceWe introduced the Structured Neural Network (StrNN), which encodes conditional independencies in data by masking pathways in the network. We integrate the StrNN into autoregressive (StrAF) and continous (StrCNF) normalizing flows to obtain structured density estimation. StrAF improves the state-of-the-art in causal inference when estimating interventional and counterfactual distributions. Neural Information Processing Systems (NeurIPS), 2023
|
|
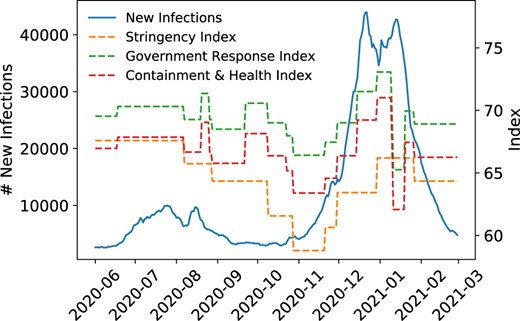
PAN-cODE: COVID-19 forecasting using Conditional Latent ODEsWe leverage the Latent ODE architecture for short-term COVID-19 caseload forecasting, conditioning latent trajectories on government interventions to generate alternative scenarios. Our approach performs comparably or better than state-of-the-art models when applied to US regions. Journal of the American Medical Informatics Association, 2022
|
|
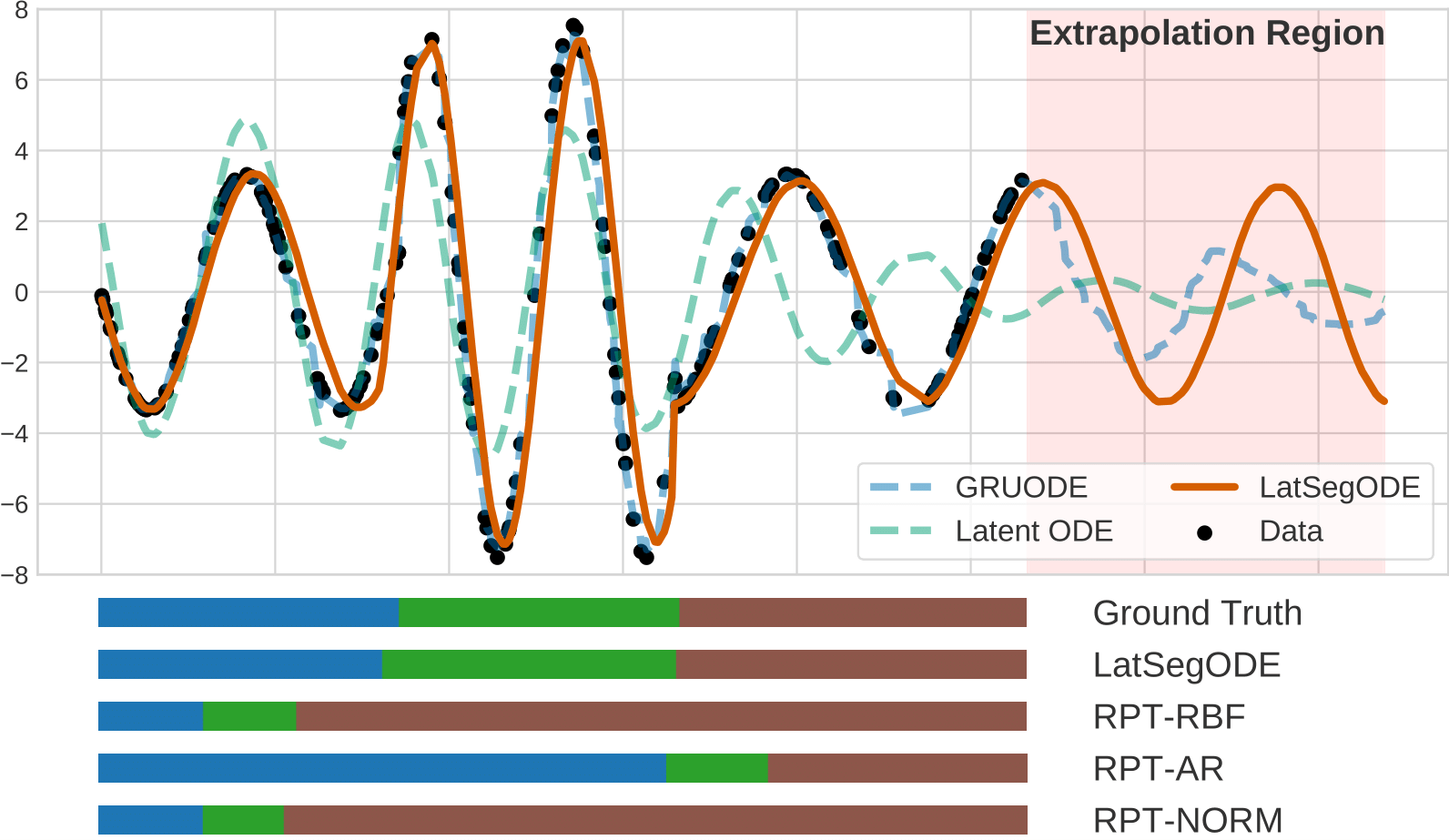
Segmenting Hybrid Trajectories using Latent ODEsWe developed LatSegODE to model hybrid trajectories with discontinuous jumps and dynamic mode changes. After fitting Latent ODEs to trajectory primitives, we apply changepoint detection to identify optimal points for restarting ODE dynamics. Reconstructions are scored using marginal likelihood to regularize the number of detected changepoints. International Conference of Machine Learning (ICML) 2021
|
|
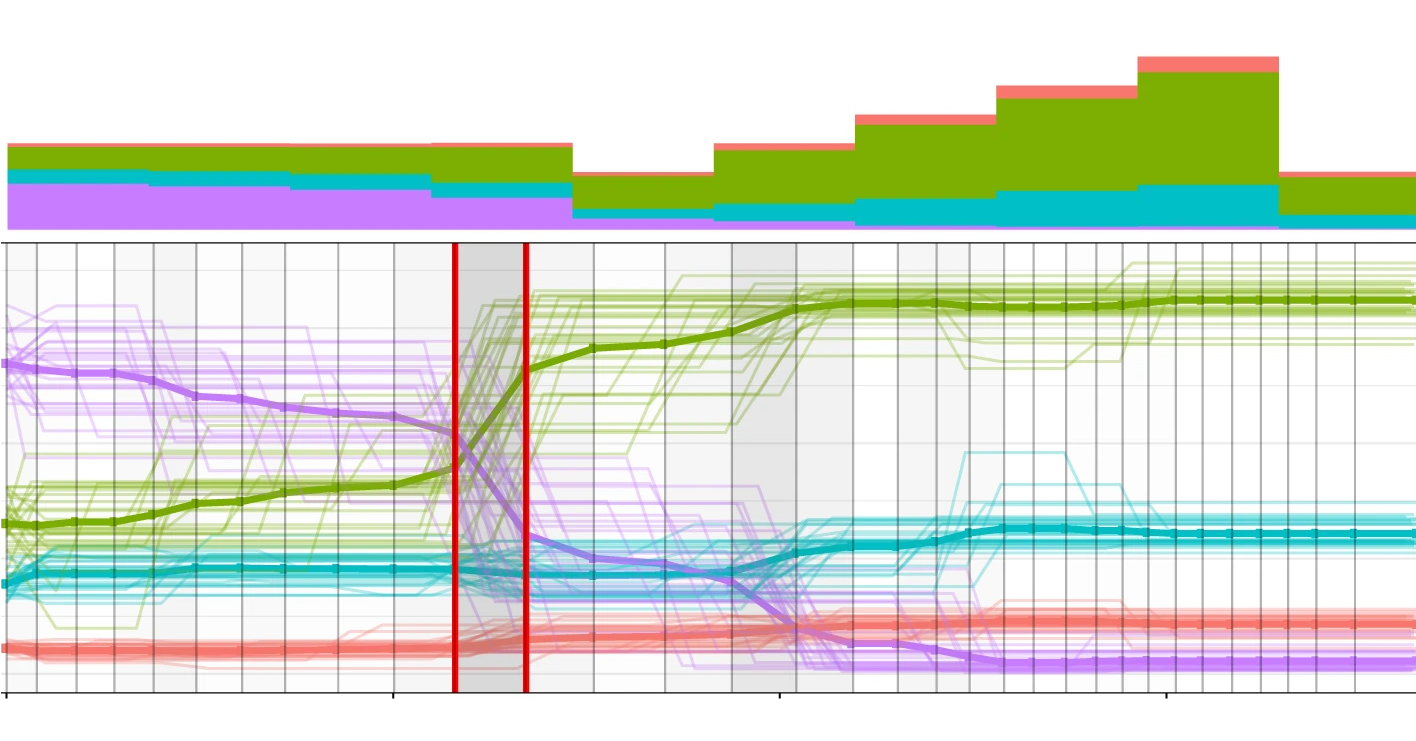
Reconstructing evolutionary trajectories of mutation signature activities in cancer using TrackSig
We developed a method to reconstruct mutational signature trajectories in cancer populations over time. Mutational activity was modeled using a mixture of multinomials, and joint optimal segmentation was applied to divide trajectories into distinct mutagenic periods. Nature Communications, 2020
|
|
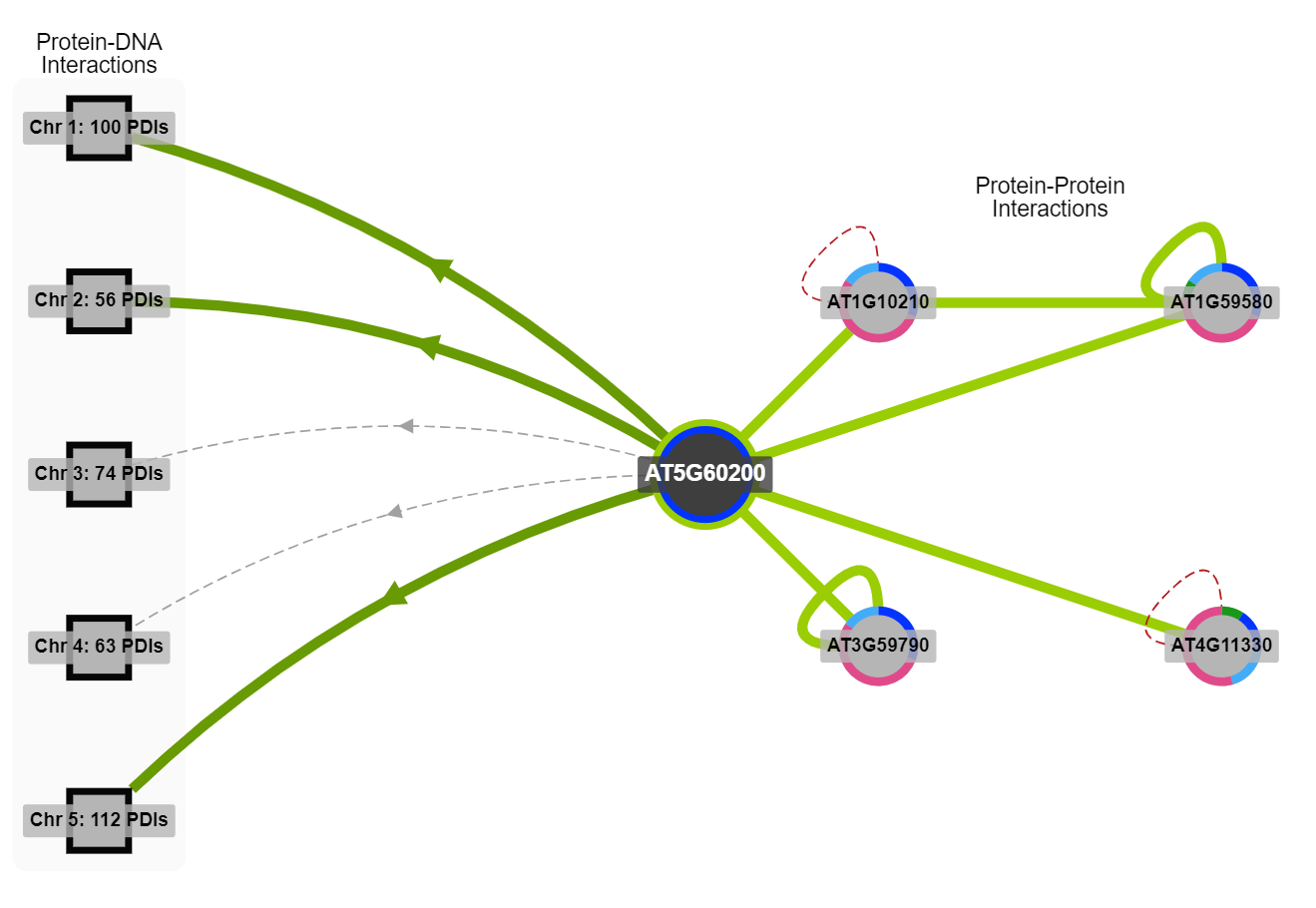
ePlant : Visualizing and Exploring Multiple Levels of Data for Hypothesis Generation in Plant BiologyWe created an analytic portal for plant model species, integrating over 12 levels of visualizations across millions of data points. I developed the Protein Interactions Viewer, which showcases known and predicted interactions between proteins and DNA. Plant Cell, 2017
|
|
Projects
Predicting Patient Reported COPD Symptoms Using Smart Device Sensor Data
We applied various classical and deep learning methods to predict the onset of symptoms of COPD exacerbation from smartwatch sensor data. Developed as part of CSC2541, with data obtained from the WearCOPD project.
Clustering Subclonal Phylogenies Using Gaussian Mixture Models
We applied clustering methods on phylogenetic tree reconstructions of cancer tumors. Non-parametric GMM clustering was applied on MCMC sample outputs to obtain characteristic phylogentic tree reconstructions from PhyloWGS.
Teaching
- CSC311: Intro to Machine Learning (2020-2023)
- CSC413: Neural Networks and Deep Learning (2020-2022)
- CSC207: Software Design (2019)